Metabolomics
Using MetaHD: A multivariate meta-analysis model for metabolomics data
by Jayamini C. Liyanage, Luke Prendergast, Robert Staudte and Alysha De Livera
MetaHD is an R package that performs multivariate meta-analysis for high-dimensional metabolomics data for integrating and collectively analysing individual-level metabolomics data generated from multiple studies as well as for combining summary estimates. This approach accounts for correlation between metabolites, considers variability within and between studies, handles missing values and uses shrinkage estimation to allow for high dimensionality. Read more →
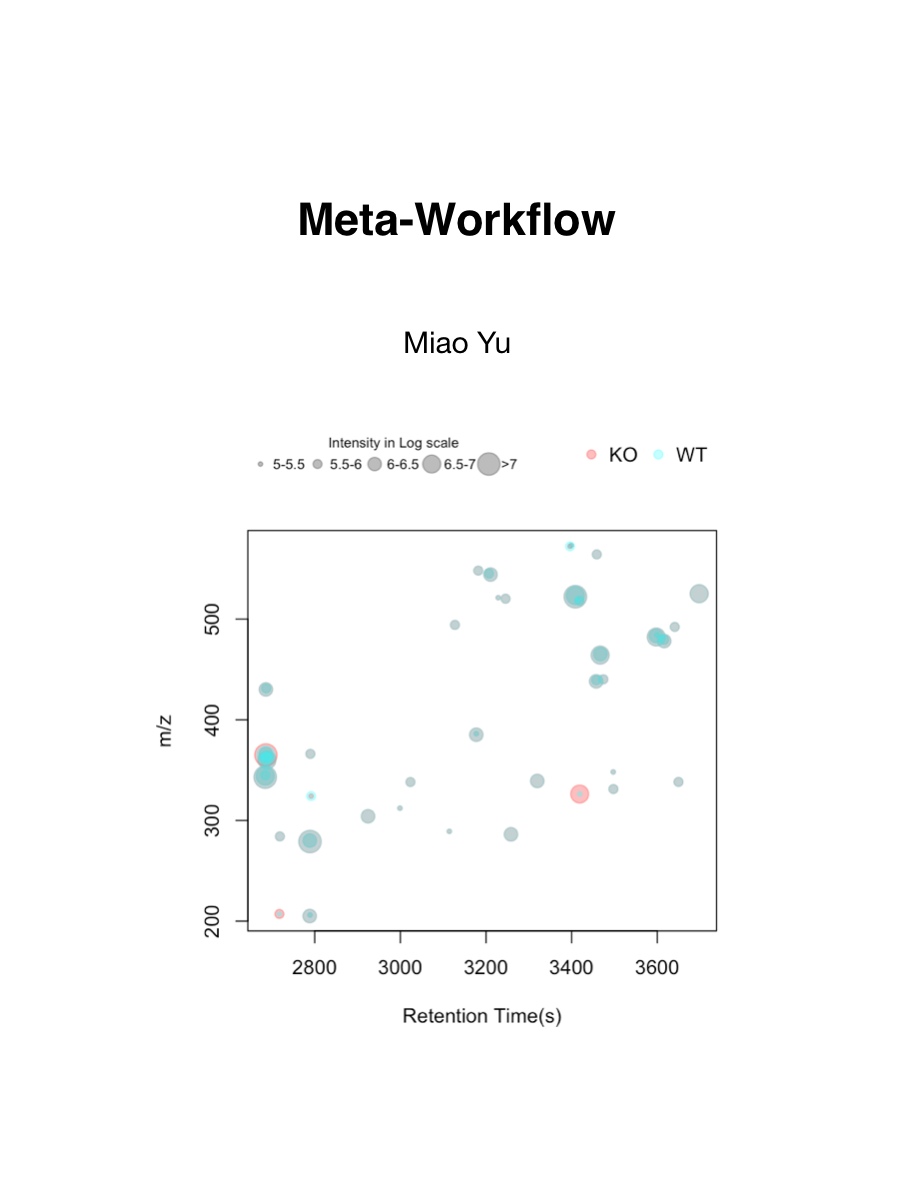
Meta-Workflow
by Miao YU
This is a workflow for metabolomics studies. […] This is an online handout for mass spectrometry based metabolomics data analysis. It would cover a full reproducible metabolomics workflow for data analysis and important topics related to metabolomics. Here is a list of topics: This is a book written in Bookdown. You could contribute it by a pull request in Github. A workshop based on this book could be found here. Meanwhile, a docker image xcmsrocker is available for metabolomics reproducible research. R and Rstudio are the software needed in this … Read more →
pctax: Analyzing Omics Data with R
by Chen Peng
Chen Peng Omics technologies have found extensive applications in biology, encompassing disciplines such as microbiome studies, transcriptomics, metabolomics, and beyond. These technologies generate diverse datasets based on distinct methodologies. Following specific upstream processing, omics data is often transformed into feature abundance tables, representing entities like genes, metabolites, or taxa. These feature abundance tables, coupled with experimental design metadata and various annotation data, serve as the foundation for downstream bioinformatics analysis and visualization, … Read more →