1 Introduction
A current challenge in Ecology is to understand the impact of the accelerated loss of biodiversity in ecosystems,(Ceballos et al. 2015) caused mainly by human activities on the planet (Novacek and Cleland 2001; Koh et al. 2004). In the framework of the Aichi 2020 targets, set up by the international community with the objective to tackle the problem of global biodiversity loss, Essential Biodiversity Variables(EBV’s) have been proposed to identify key measures for adequately monitor and predict future biodiversity scenarios(Pereira 2013 ; Kissling et al. 2015). In this context, the availability of biodiversity data(e.g. presence - absence, abundance, species interactions, etc.) across relevant spatial and temporal scales is one of many challenges for the global implementation of EBV’s(Kissling et al. 2015).
Global biodiversity research depends on high - level concepts abstracted and put together to synthesize a variety of information independently produced by local observers, teams, and institutions working differently all over the world(Bisby 2000). This array of sources creates an intrinsic difficulty on “knowing what is where” and “comparing like with like”(Bisby 2000), fueling the need for integration of different ecological data and information over various geographical and environmental scales(Thuiller et al. 2013; Kissling and Schleuning 2015; Poissot et al. 2016). The relatively recent emerged field of biodiversity informatics has already started to deliver profound advances with the objective of turning the world wide web into a “giant biodiversity information system”(Bisby 2000). Hence, there is a growing need for optimizing the access to available knowledge on Ecology(Pilsk et al. 2016; Lyal 2016).
Existing data on worldwide biodiversity is scattered around many databases, print on paper, or published in many media formats with and without interactive searching capabilities (Edwards et al. 2000; Lyal 2016). Open availability of biodiversity data foster opportunities for collaboration between researchers (GBIF 2016) and promotes the discoverability of a “big - picture” on biodiversity around the globe. Data Integration projects like the the Global Biodiversity Information Facility(GBIF) for species occurrence data and TRY database for species traits are very good examples of integrating data to making it available to a wider audience of ecologists and public in general (Edwards et al. 2000; Proença et al. 2016). Many databases can now be accessed in a programmatic way(Poissot et al. 2016) as computational tools have been developed to efficiently query and download data (e.g Geocat, rGbif, Flickrr). However, given the scope of the EBV’s, the demands for datasets at multiple spatial and temporal scales is far from being met (Proença et al. 2016).
In addition to ecological research on species distributions, we see a growing interest for the study of biotic interactions across trophic levels, partially in the context of climate change (Bascompte & Jordano 2007; Araujo et al. 2007; Tylianakis et al. 2008; Kissling & Schleuning 2015). Under climate change, shifts in host plant distribution ranges can influence the abundance of their herbivores or pollinators, becoming an important driver of the selection process(Van der Putten et al. 2010). However, species distributions models and predictions on climate–induced habitat range shifts rarely take biotic interactions into account (Kissling et al. 2010) which limits the ability of such models to understand the global outcomes of processes like rapid shifts in species distributions (Betzholtz et al. 2013) and to relate them to process and patterns like species co-extinctions (Koh et al. 2004) and biological homogenization (Olden 2006).
Macroecological questions on biotic interactions have been typically addressed as comparative research studies of local networks across large geographical scales (Dalsgaard et al. 2011; 2013, Schleuning et al. 2012; Trøjelsgaard and Olesen 2013; Martín Gonzales et al. 2015). Monetary, human and time constraints has restrained data collection on species interactions to be limited to local scales (Poissot et al. 2016). Thus, comparative macroecological studies rely on information limited by sampling. Data from local field studies, citizen science, satellite and remote sensing data have been recently used as aggregated datasets to study trends on species distributions(van Swaay et al., 2013; Flombaum et al. 2013; Hudson et al. 2014). Moreover, “synthetic” datasets (Poissot et al. 2016) have been proposed as a novel, rapid and cost - effective alternative to answering macroecological questions on multi - species interactions by aggregating species interactions information hosted in databases freely available on the web (Figure 1.1).
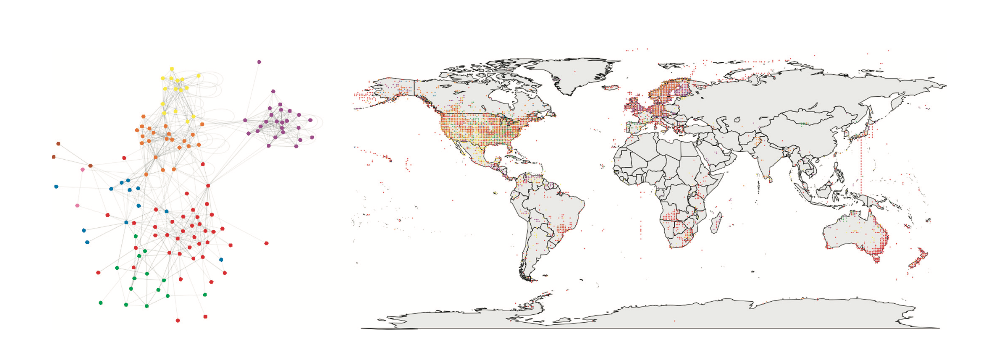
Figure 1.1: Example of the use of synthetic datasets. Left: Food web with data on species interactions from GLOBI, colored by modules(clusters of highly interacting nodes). Right: Ocurrence data(from BISON and GBIF) of species included in the foodweb, also colored by module. Figure obtained from Poissot et al.(2016)
Ecological information on species interactions can be located in specialized biodiversity data repositories such as the Interaction Web Database (IWDB) and Global Biotic Interactions (GloBI). Nevertheless, the information available in those sources is still incomplete and its growth currently depends solely on datasets contributions from independent researchers. Much of ecological records on species interactions are still over-dispersed over the internet, placed in local and global scholarly sources(e.g. Global: Scopus, Web of Science, e.g. Local: Biblat); university repositories (Gamboa 2012; Reyna Espinosa 2015) and non – scholarly sources (e.g. Flickr, bioversity webpages, online books, etc.)(Shanmughavel 2007; Barve 2014; Miranda & Strüssmann 2016). Additionally, the current body of published academic literature in Ecology holds a good volume of ecological observations on species interactions mostly product of unrelated localized studies (Beck 2006, Proença et al. 2016).
To generate new hypothesis in the search for large-scale patterns of species interactions (Poissot et al. 2016) and to develop new representative EBV’s datasets (Proença et al. 2016), ecological observations on species interactions can be extracted from the content of published literature (Poissot et al. 2016; Lyal 2016). This can be done with manual literature surveys (Strong and Leroux 2014), but when considering a large number of taxa, this process becomes daunting (Poissot et al. 2016). Machine learning and text mining techniques, such as Abductive ILP (A/ILP), have explored to automatically identify and infer trophic links from literature (Bohan and Tamaddoni-Nezhad 2011, Milani et al. 2012, Tamaddoni-Nezhad et al. 2014) with promising initial results. However, all future developments in this field are currently limited as not all major publisher houses allows researchers to mine their own created literature (Poissot et al. 2016).
In this literature review, using computational tools developed for text mining, I will evaluate:
- Different ways to automatically (or semi - automatically) search and download ecological literature;
- the performance of different methods for automatic classification of articles with mentions of species interactions;
- The use of automatic extraction of specific text features(such as species names and geographical location) to discriminate articles of interest and
- the use of customized article summaries to improve the reading time of large portions of literature. With this processes, I aim to build a framework that helps ecological researchers interested macroecology to search and identify potential articles of interest containing records on species interactions from the pool of published literature in a programmatic way.
For the purpose of this literature review, I will focus on computational tools that are open and (mostly) free; as I believe in their importance to foster the qualitative development of science around the globe. In addition, as a practical example, I’ll focus on frugivory plant - animal interactions and use resources written in the R language. Nevertheless, most if not all of the applications I will mention in this document have their counterparts written in other computer languages such as python, Perl, etc.