Chapter 2 Exploratory Analysis
2.1 Proportions
library(tidyverse)
library(viridis)
library(colorspace)
t1 %>%
mutate(mm = ifelse(resultado_micro==1, "micro+", "micro-"),
ss = ifelse(SEROPOSITIVE=="Positive", "sero+", "sero-"),
out = paste0(mm, " | ",ss)) %>%
filter(!is.na(resultado_micro)) %>%
ggplot(aes(x = age_cat, fill = out)) +
geom_bar(position = "fill") +
scale_fill_discrete_sequential(palette="ag_Sunset") +
labs(y = "proportion", x = "Age category", fill = "Outcome") +
theme_bw() +
facet_grid(~nm_sex, labeller = labeller(nm_sex = c("0_female" = "Female", "1_male" = "Male")))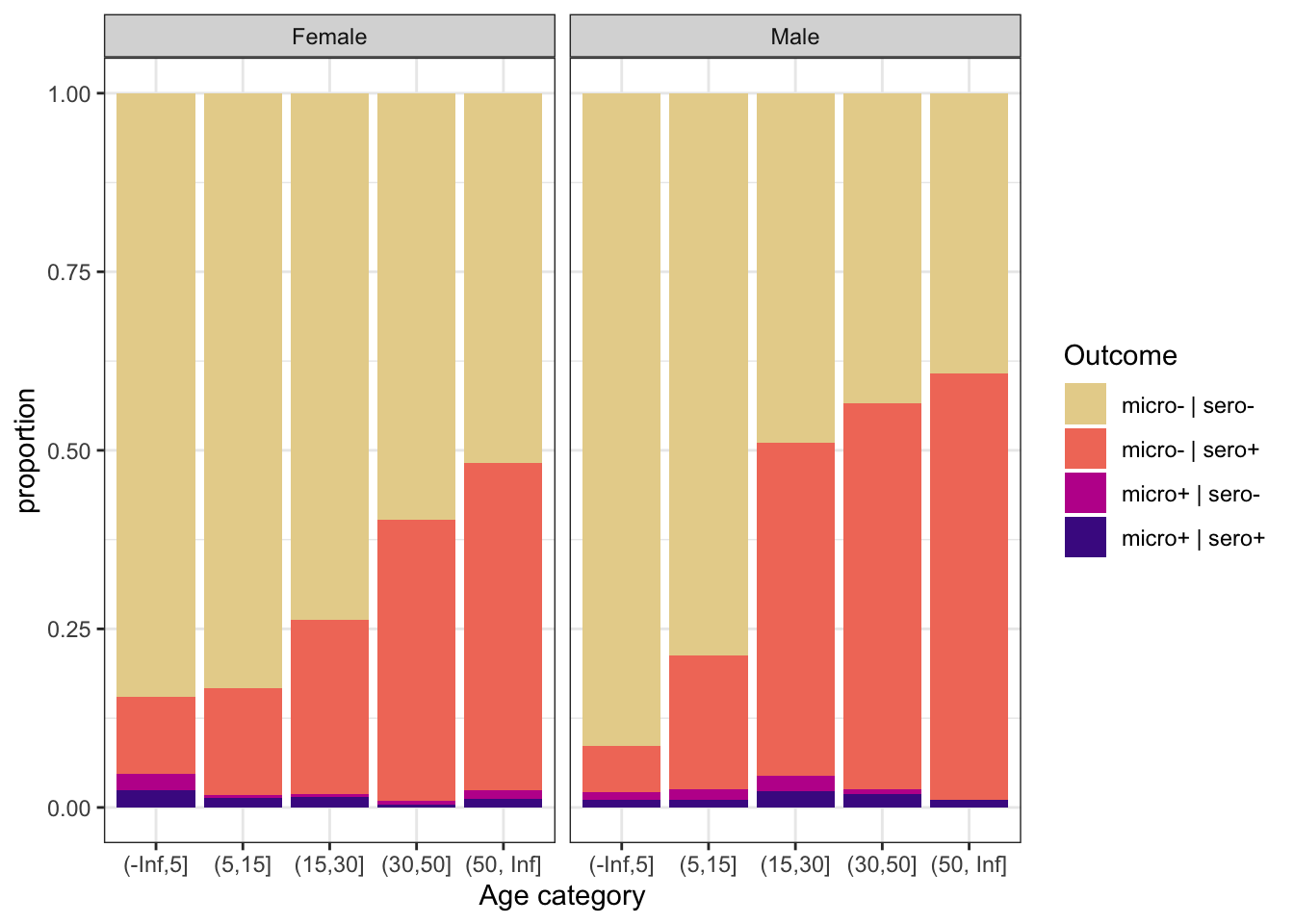
t1 %>%
mutate(mm = ifelse(resultado_micro==1, "micro+", "micro-"),
ss = ifelse(SEROPOSITIVE=="Positive", "sero+", "sero-"),
out = paste0(mm, " | ",ss)) %>%
filter(!is.na(resultado_micro)) %>%
ggplot(aes(x = age_cat, fill = out)) +
geom_bar(position = "fill") +
scale_fill_discrete_sequential(palette="ag_Sunset") +
labs(y = "proportion", x = "Age category", fill = "Outcome") +
theme_bw() +
facet_grid(~fever, labeller = labeller(fever = c("0" = "No Fever", "1" = "Fever")))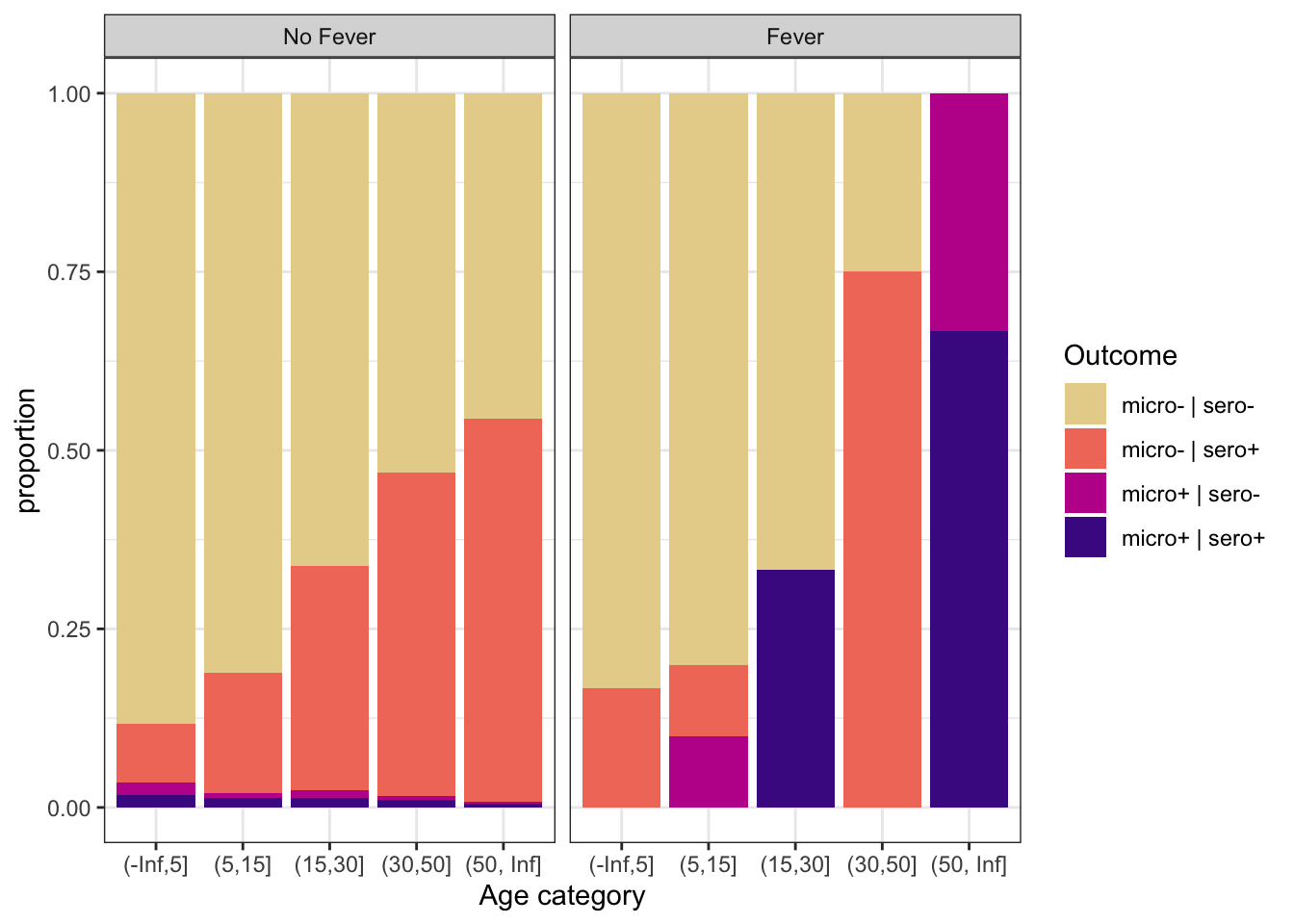
t1 %>%
mutate(mm = ifelse(resultado_micro==1, "micro+", "micro-"),
ss = ifelse(SEROPOSITIVE=="Positive", "sero+", "sero-"),
out = paste0(mm, " | ",ss)) %>%
filter(!is.na(resultado_micro)) %>%
ggplot(aes(x = age_cat, fill = out)) +
geom_bar(position = "fill") +
scale_fill_discrete_sequential(palette="ag_Sunset") +
labs(y = "proportion", x = "Age category", fill = "Outcome") +
theme_bw() +
facet_grid(~area, labeller = labeller(area = c("0_periurban" = "periurban", "1_rural" = "rural")))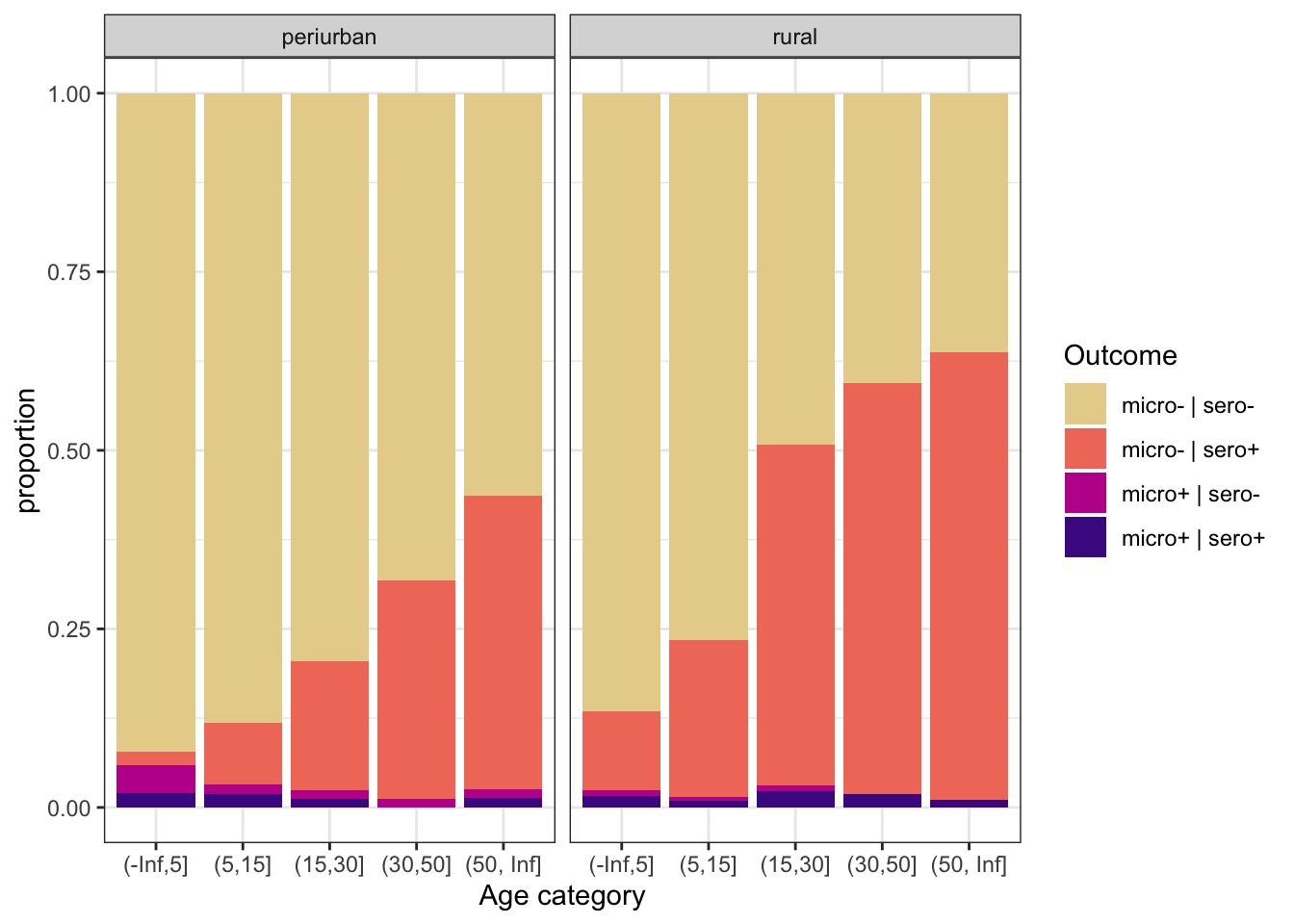
2.2 Maps
2.3 Distance plot
2.3.1 Reference Febrile case
library(magrittr)
library(stringr)
d3 <- d2 %>%
filter(fever==1 | hist_fever==1) %>%
distinct(id_house, .keep_all = T)
m <- st_distance(d2,d3) %>%
as.data.frame() %>%
set_colnames(d3$id_house) %>%
mutate(id_study = d2$id_study,
id_muestra = d2$id_muestra) %>%
relocate(id_study, id_muestra) %>%
gather(target,distance_m,-id_study, -id_muestra) %>%
mutate(distance_m = as.numeric(distance_m),
area_o = str_sub(id_study,1,3),
area_t = str_sub(target,1,3)) %>%
group_by(id_study, id_muestra, area_o, area_t) %>%
summarise(min_dist_m = min(distance_m, na.rm = T)) %>%
filter(area_o==area_t)
d4 <- d2 %>%
inner_join(m, by="id_muestra")
d4 %>%
mutate(dist_cat = cut(min_dist_m, breaks = c(-Inf,10,20,30,40,Inf))) %>%
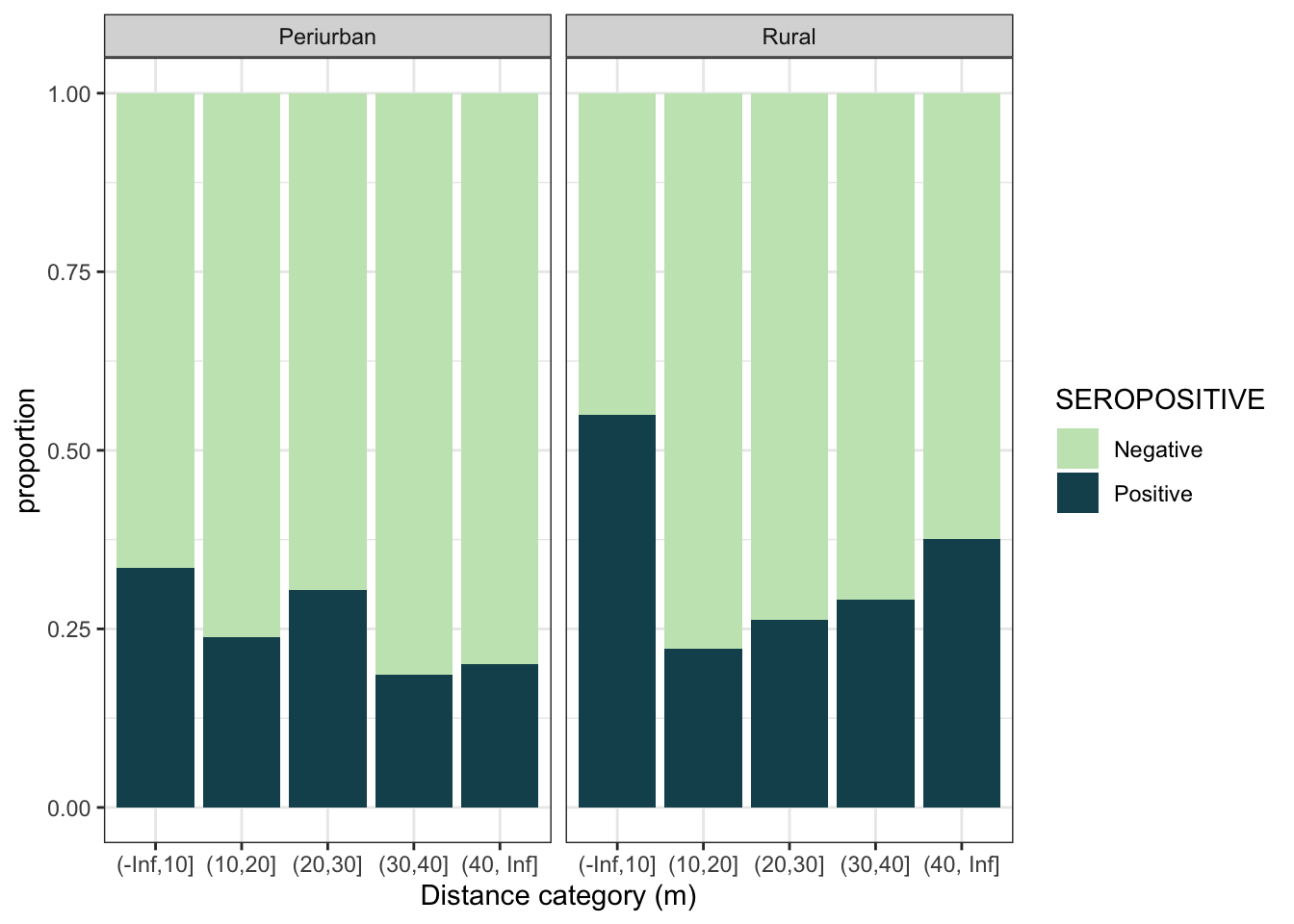
ggplot(aes(x = dist_cat, fill = SEROPOSITIVE)) +
geom_bar(position = "fill") +
scale_fill_discrete_sequential(palette="BluGrn") +
labs(y = "proportion", x = "Distance category (m)") +
theme_bw() +
facet_grid(~area, labeller = labeller(area = c("0_periurban" = "Periurban", "1_rural" = "Rural")))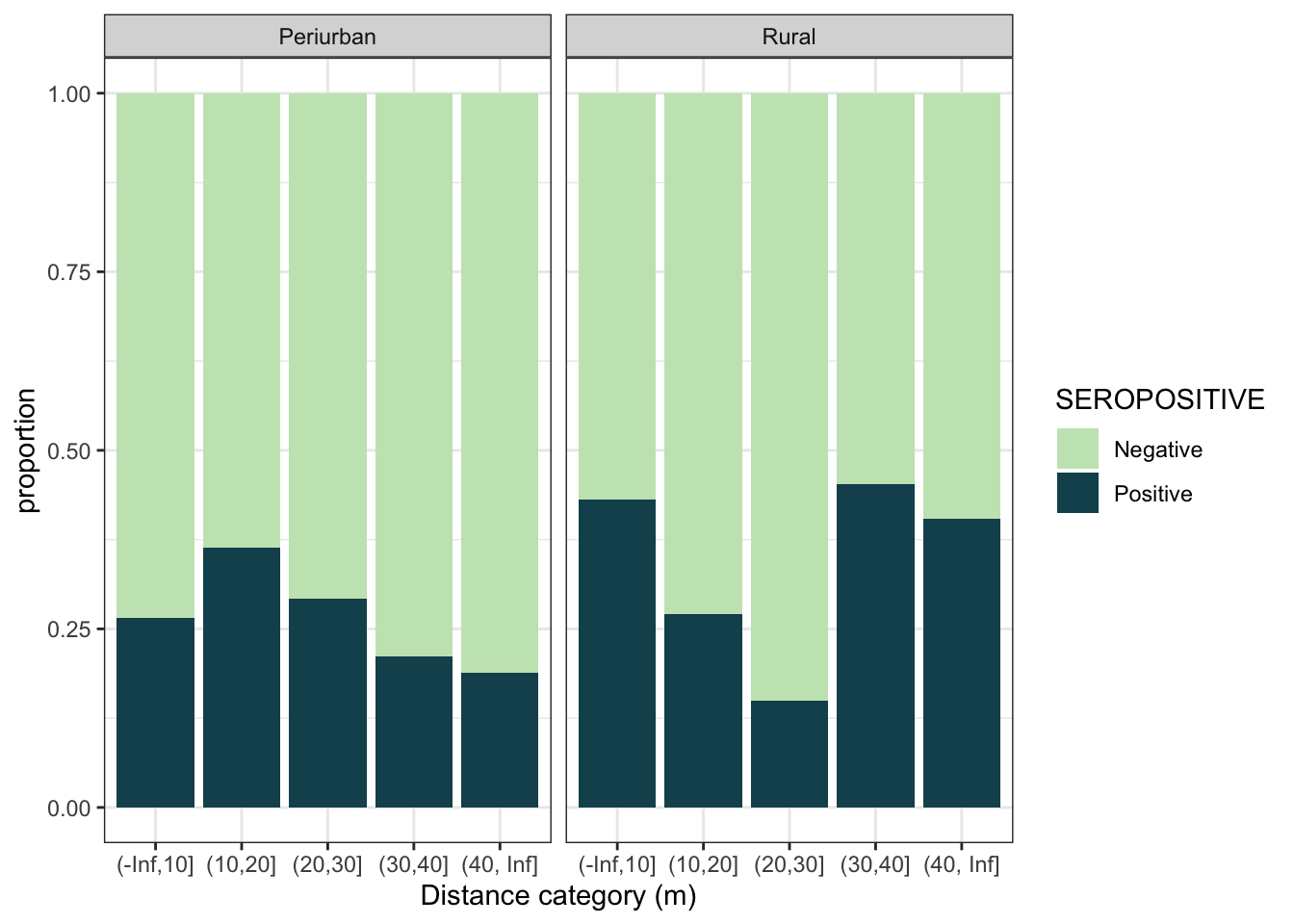
2.3.2 Reference Seropositive case
library(magrittr)
library(stringr)
d3 <- d2 %>%
filter(sero==1) %>%
distinct(id_house, .keep_all = T)
m <- st_distance(d2,d3) %>%
as.data.frame() %>%
set_colnames(d3$id_house) %>%
mutate(id_study = d2$id_study,
id_muestra = d2$id_muestra) %>%
relocate(id_study, id_muestra) %>%
gather(target,distance_m,-id_study, -id_muestra) %>%
mutate(distance_m = as.numeric(distance_m),
area_o = str_sub(id_study,1,3),
area_t = str_sub(target,1,3)) %>%
group_by(id_study, id_muestra, area_o, area_t) %>%
summarise(min_dist_m = min(distance_m, na.rm = T)) %>%
filter(area_o==area_t)
d4 <- d2 %>%
inner_join(m, by="id_muestra")
d4 %>%
mutate(dist_cat = cut(min_dist_m, breaks = c(-Inf,10,20,30,40,Inf))) %>%
ggplot(aes(x = dist_cat, fill = SEROPOSITIVE)) +
geom_bar(position = "fill") +
scale_fill_discrete_sequential(palette="BluGrn") +
labs(y = "proportion", x = "Distance category (m)") +
theme_bw() +
facet_grid(~area, labeller = labeller(area = c("0_periurban" = "Periurban", "1_rural" = "Rural")))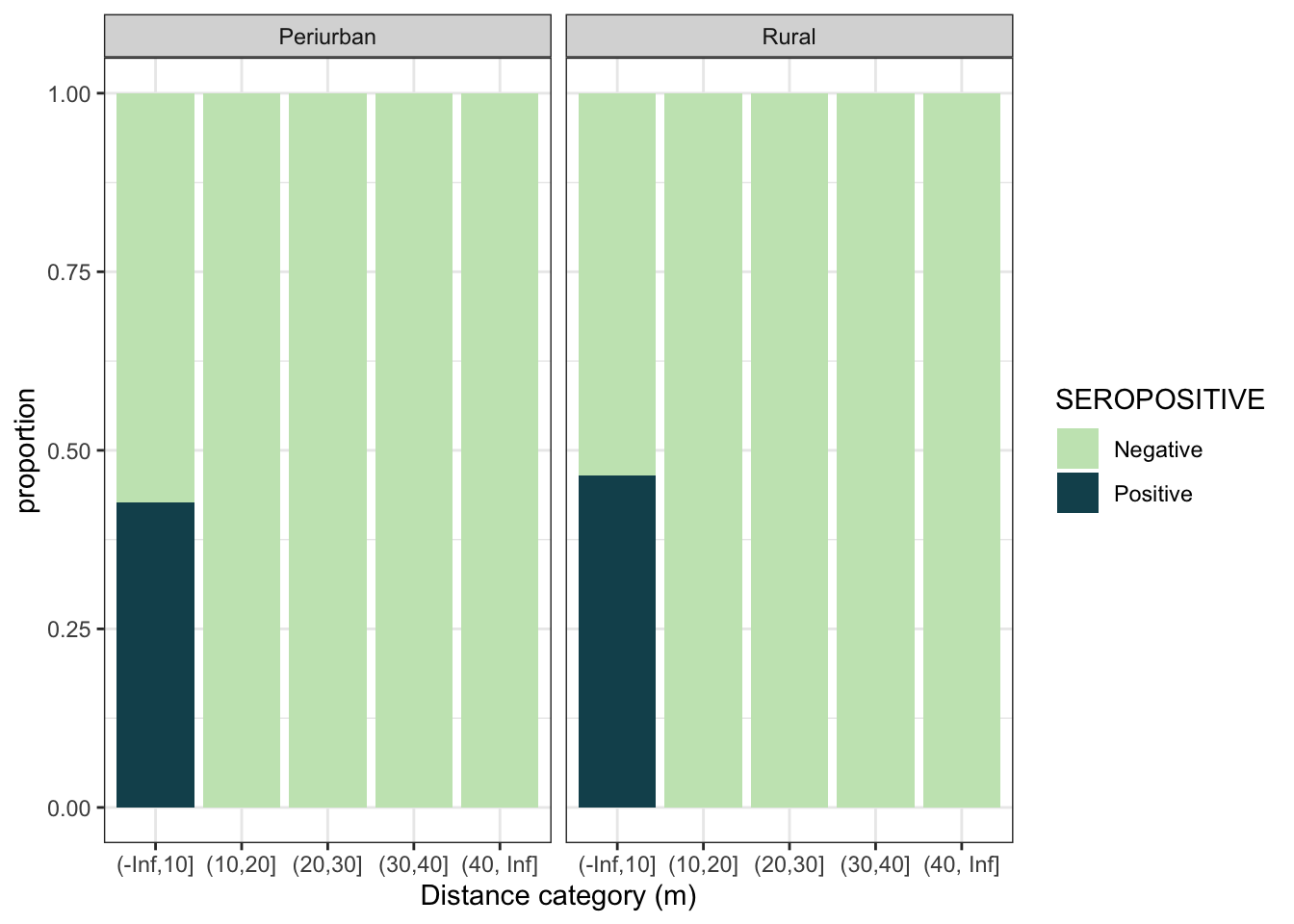
2.3.3 Reference PCR Positive
library(magrittr)
library(stringr)
d3 <- d2 %>%
filter(pcr_pos==1) %>%
distinct(id_house, .keep_all = T)
m <- st_distance(d2,d3) %>%
as.data.frame() %>%
set_colnames(d3$id_house) %>%
mutate(id_study = d2$id_study,
id_muestra = d2$id_muestra) %>%
relocate(id_study, id_muestra) %>%
gather(target,distance_m,-id_study, -id_muestra) %>%
mutate(distance_m = as.numeric(distance_m),
area_o = str_sub(id_study,1,3),
area_t = str_sub(target,1,3)) %>%
group_by(id_study, id_muestra, area_o, area_t) %>%
summarise(min_dist_m = min(distance_m, na.rm = T)) %>%
filter(area_o==area_t)
d4 <- d2 %>%
inner_join(m, by="id_muestra")
d4 %>%
mutate(dist_cat = cut(min_dist_m, breaks = c(-Inf,10,20,30,40,Inf))) %>%
ggplot(aes(x = dist_cat, fill = SEROPOSITIVE)) +
geom_bar(position = "fill") +
scale_fill_discrete_sequential(palette="BluGrn") +
labs(y = "proportion", x = "Distance category (m)") +
theme_bw() +
facet_grid(~area, labeller = labeller(area = c("0_periurban" = "Periurban", "1_rural" = "Rural")))