Chapter 3 Certainty of absence
This tool is useful if you have already conducted a green crab monitoring assessment, did not detect green crab, and want to know how certain you can be that green crab are truly absent. More specifically, given your monitoring effort, this tool finds the true capture rates that would produce no detections.
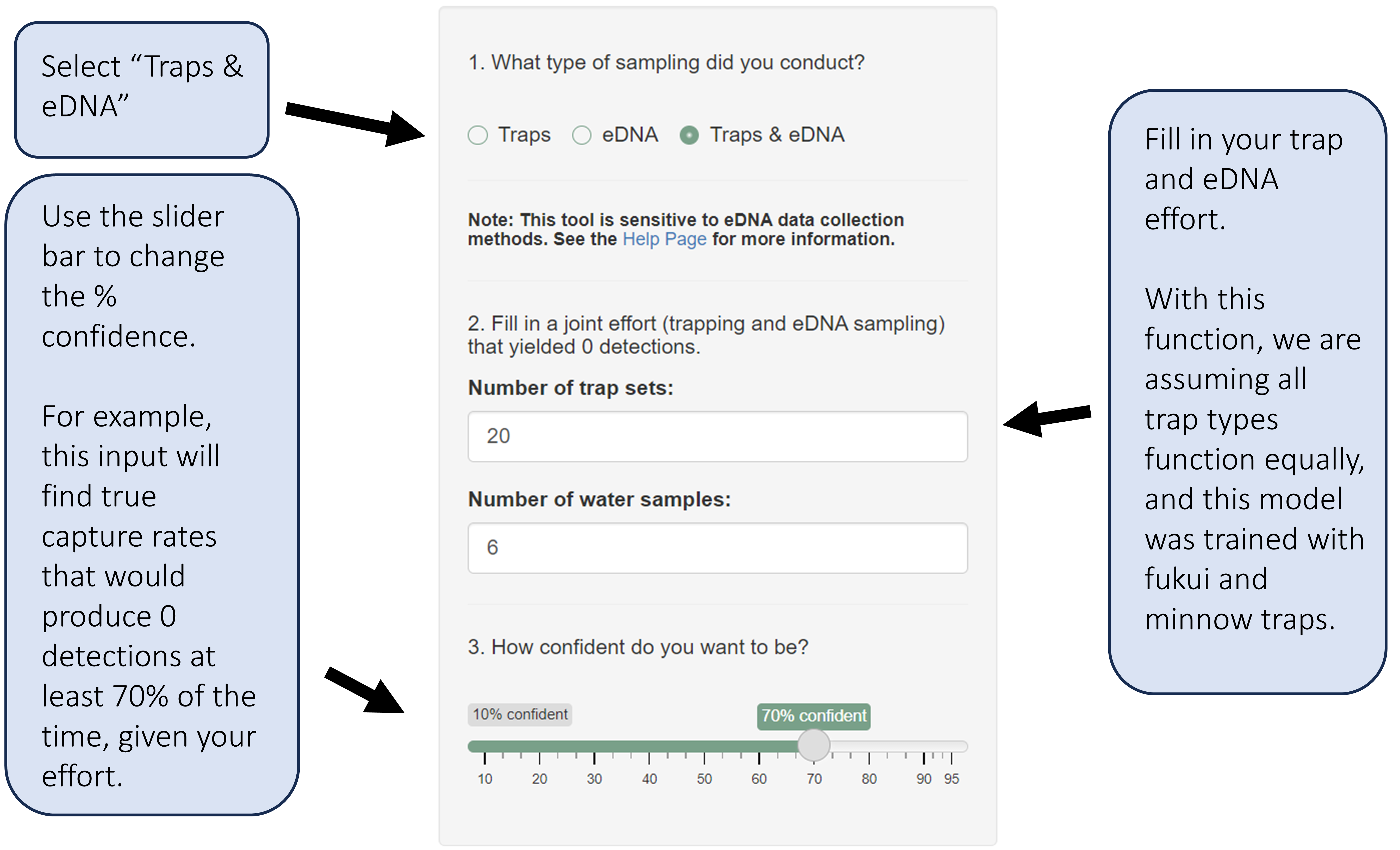
In these sections, we will focus on only trapping efforts. Refer to section 3.4 for more information about using certainty of absence with eDNA.
Note: This tool can be used to help interpret monitoring results at a site over a relatively short time period. (See section 1.3 for more info).
3.1 Inputs
Navigate to Assessment Interpretation > Certainty of Absence.
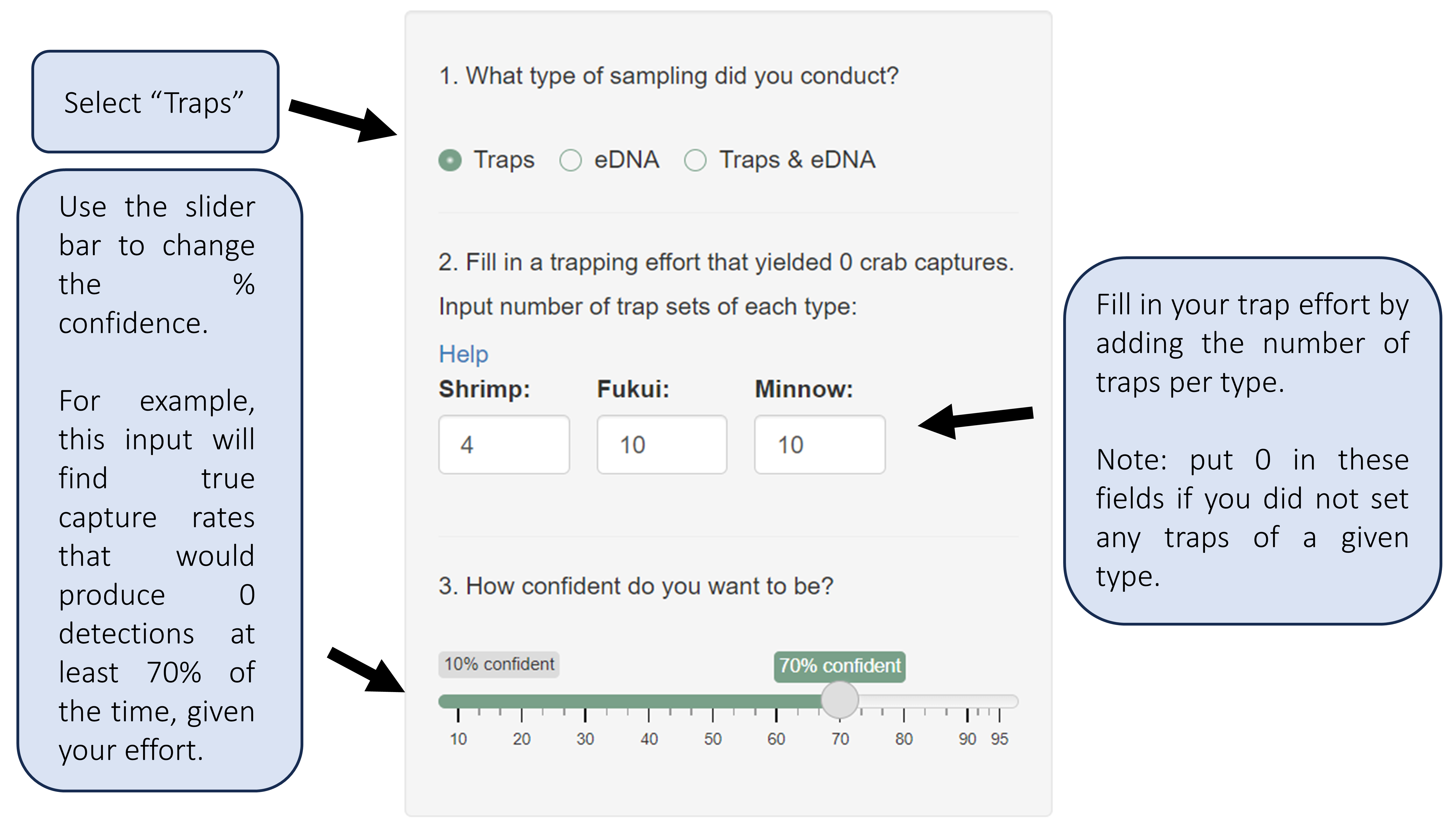
Fill in your desired inputs.
The types of traps used in this section are:
Square fukui fish traps (1.27 cm mesh)
Modified steel minnow traps (2 inch opening, 0.635 cm mesh)
Non-collapsible shrimp traps (1 inch mesh)
If the type of trap you use is not listed here, choose the trap above that functions most similarly to your trap, or contact us at crabteam@uw.edu if you have further questions. Note that one trap set refers to a soak window with one overnight tide (typically a ~16-18 hr time period).
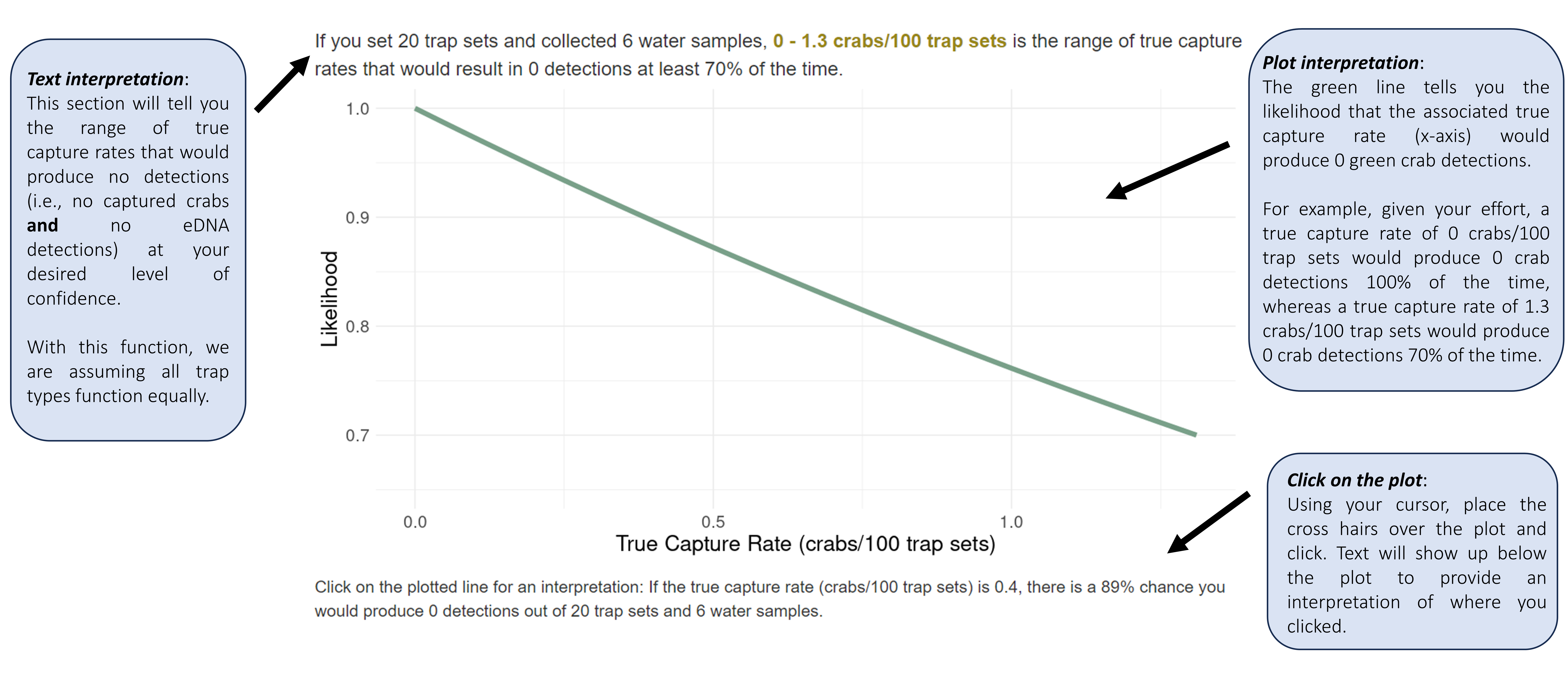
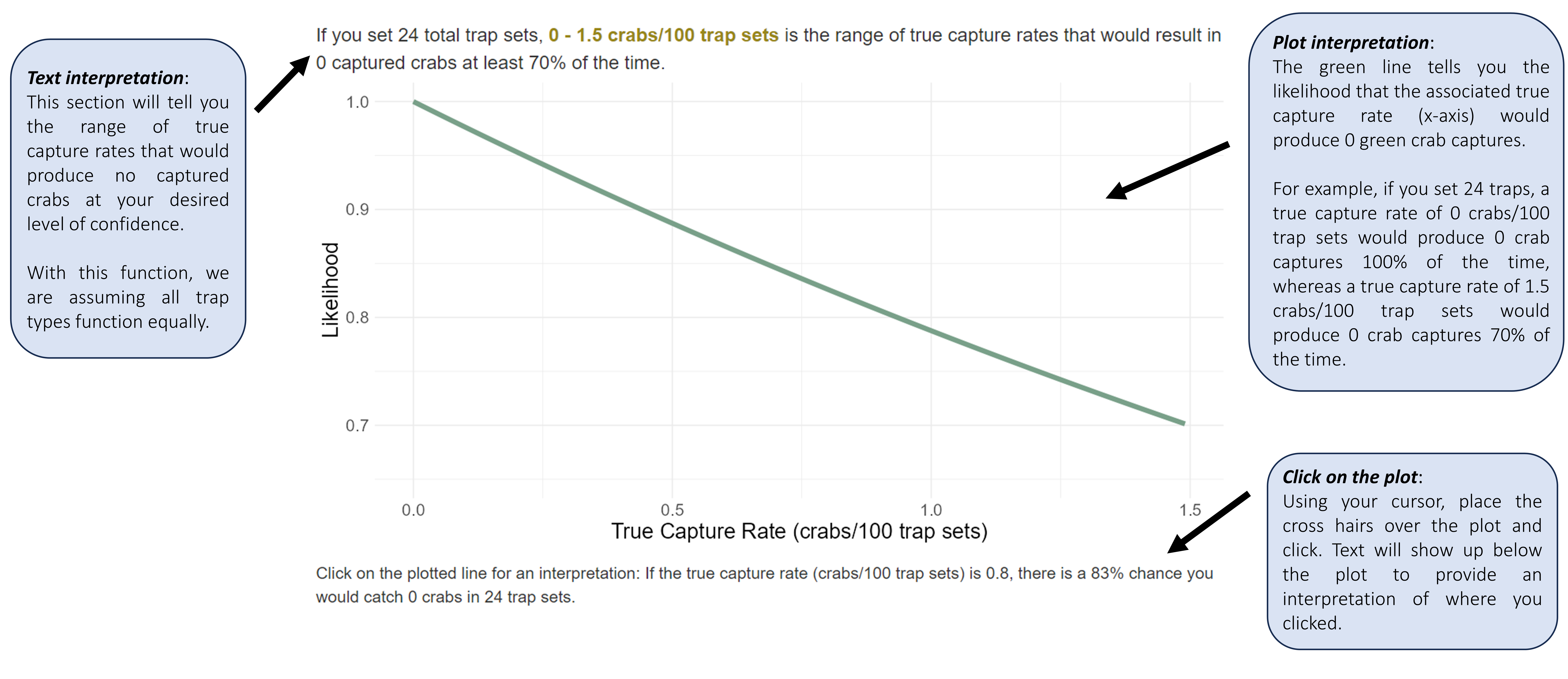
3.2 Interpreting the plot
With the choice of 70% confidence, interpret the outputs and plot.
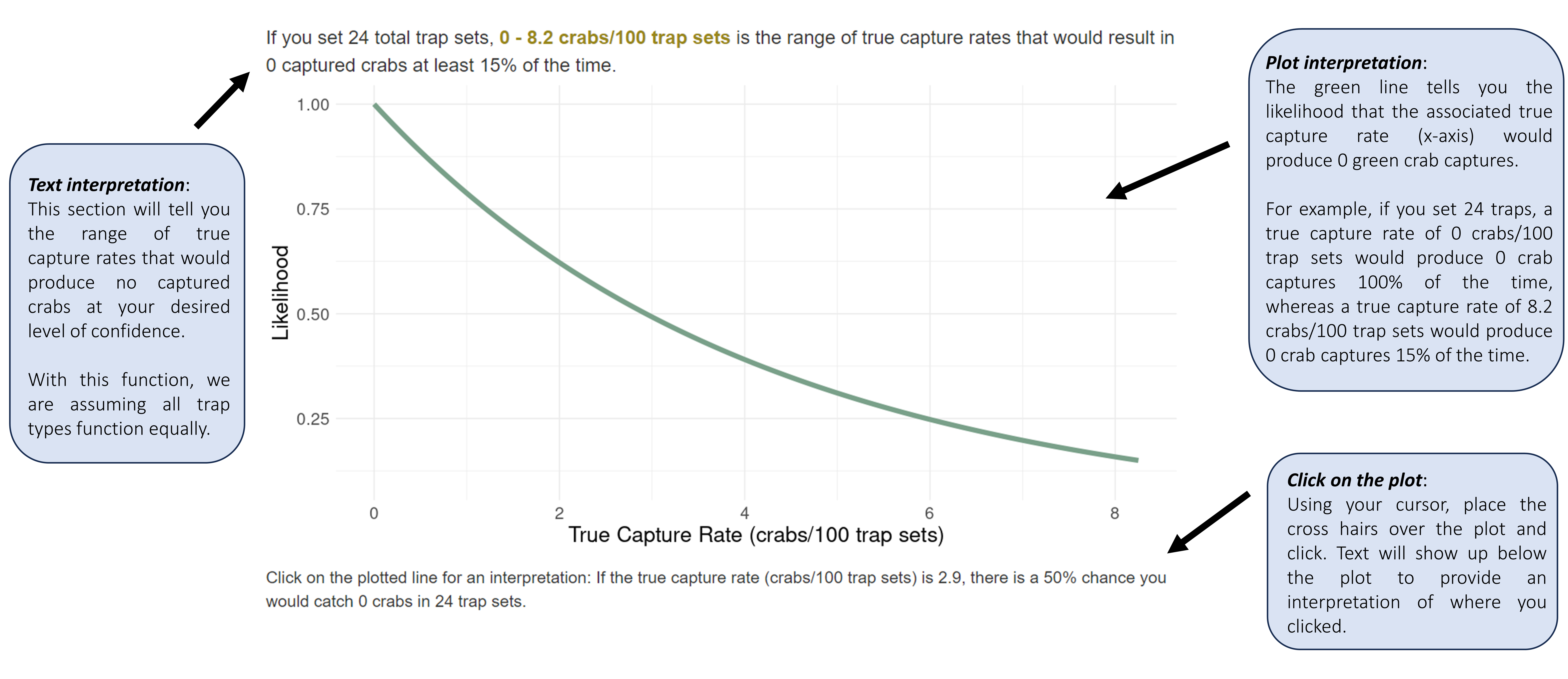
Now let’s lower the confidence to 15% and see how this changes the results.
3.3 Trap-adjusted capture rate
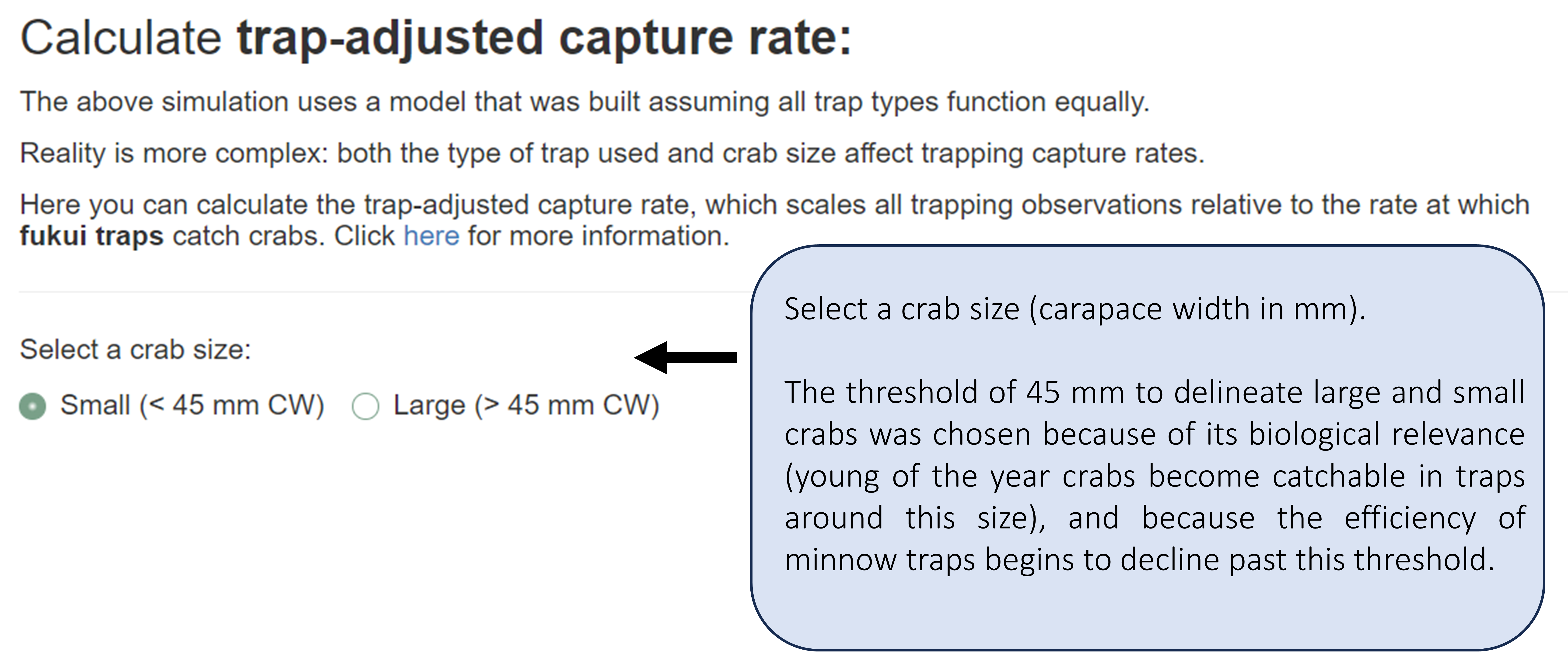
The previous section assumed all trap types function equally. However, both the type of trap and crab size affect capture rates.
In this section, you can adjust for trap type by using the trap-adjusted capture rate. This rate scales all trapping observations relative to the rate at which fukui traps catch crabs. Head to section 1.2.2 for more information about the trap-adjusted capture rate.
Here, our previous trap number inputs still apply.
Fill in the input for trap-adjusted capture rate. We’ll begin with small crabs.
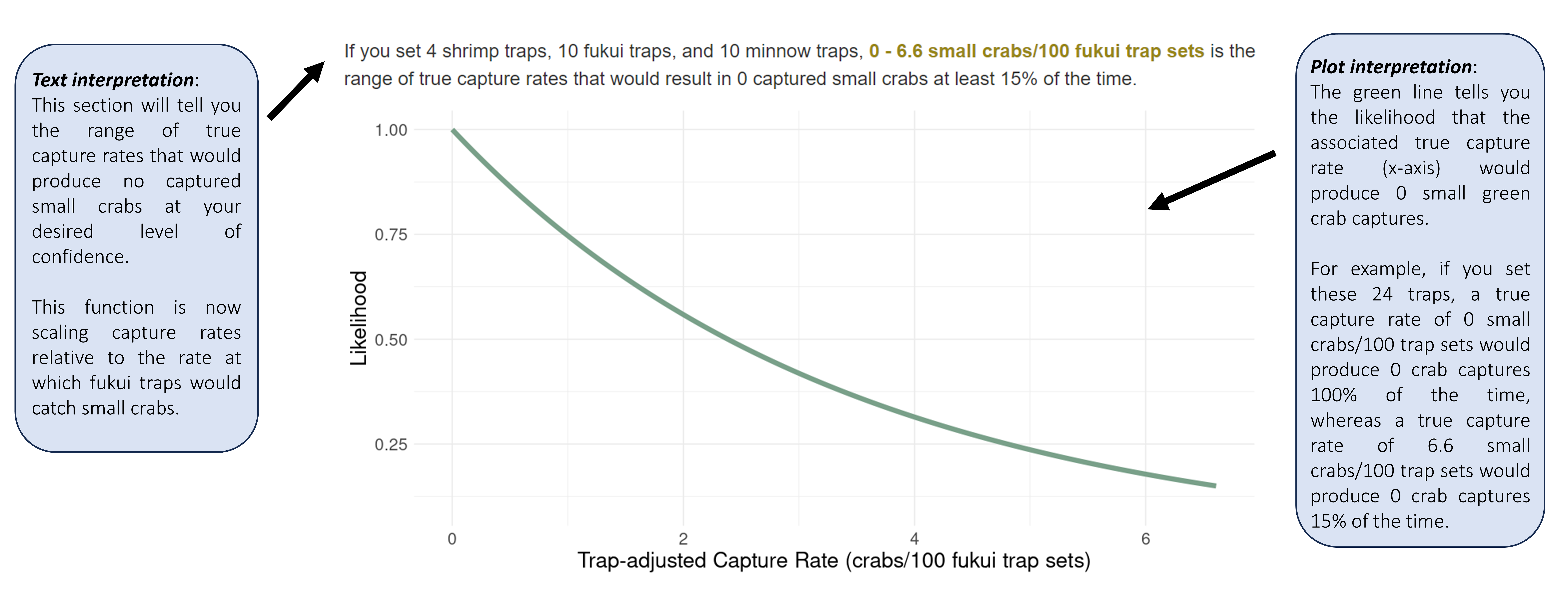
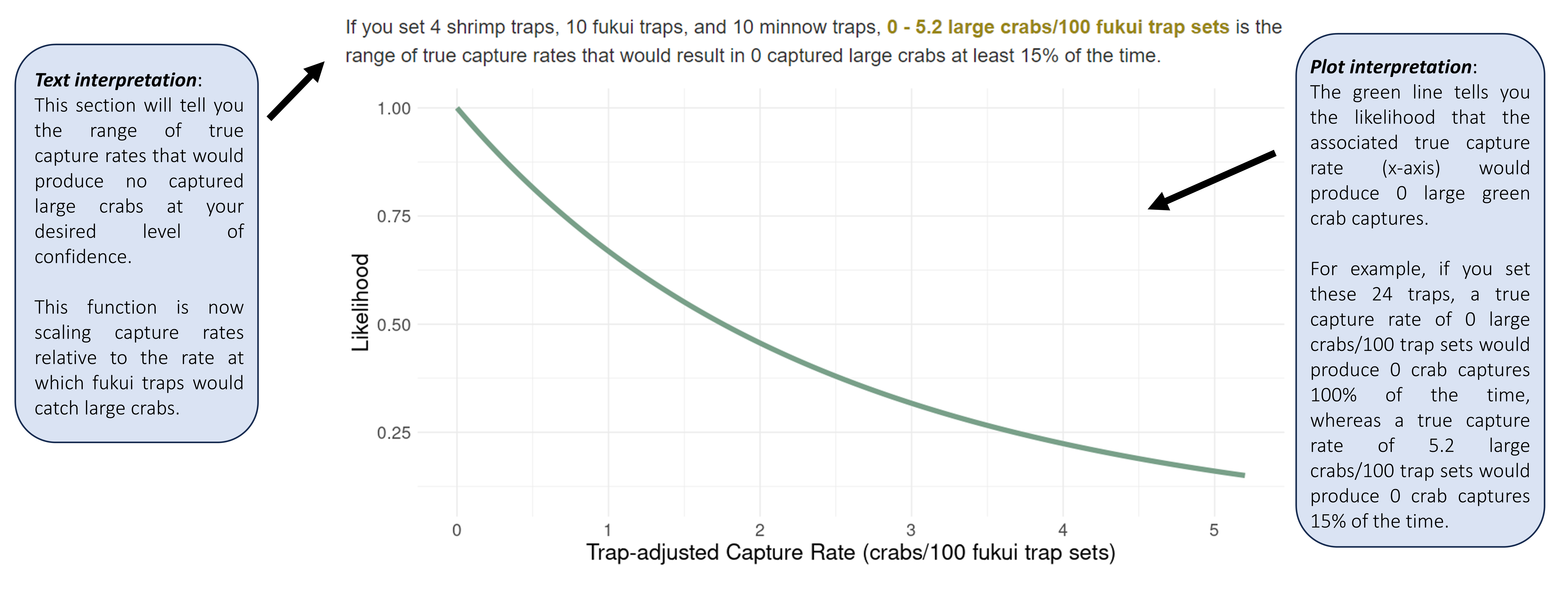
Interpret the text and plot to identify the range of trap-adjusted capture rates that would have produced 0 small crab captures, given the number and type of traps you used.
Switch the input to “large crabs” to identify the range of trap-adjusted capture rates that would have produced 0 large crab captures, given the number and type of traps you used.
3.4 Environmental DNA (eDNA)
This section of the web app is sensitive to eDNA data collection methods. The model underlying this tool was built using eDNA data collected following the protocol in Keller et al. 2022. Water samples collected for eDNA analysis using different protocols may make inference using this web tool less applicable.
The sensitivity of eDNA detection relative to trapping likely depends upon the field collection protocols (i.e. water volume, collection depth) and laboratory protocols (i.e. DNA extraction and purification methods).
Seasonality may also play a role in eDNA sensitivity. Season-specific factors like green crab activity, movement, and molting cycles can affect the rate at which organisms shed eDNA. Also, the presence and density of organic matter in the water may vary seasonally and affect eDNA extraction and PCR inhibition.
However, this tool can be useful if your eDNA protocols are similar those outlined in Keller et al. or if you want to explore how the sensitivity of eDNA samples could potentially compare to trapping.
3.4.1 Inputs
Fill in your desired inputs.
Note that one water sample refers one biological replicate with three technical replicates (i.e., qPCR reactions). Refer to Keller et al. 2022 for more information.