Chapter 3 Prediction and Inference
In the previous chapter, we have seen how to fit a GLM to a dataset and estimate its parameters \(\hat{\boldsymbol{\beta}}\). We will now learn how to make predictions, compute confidence intervals and confidence regions, and perform hypothesis tests with a fitted model.
3.1 Prediction and Confidence Intervals
Assume a GLM has been fitted, yielding \(\hat{\boldsymbol{\beta}}\in{\mathbb R}^{p}\). If we are given a new predictor vector \(\boldsymbol{x}_0\), we can compute
\[ \hat{\eta}_0 = \hat{\boldsymbol{\beta}}^T \boldsymbol{x}_0 \] and predict the response for \(\boldsymbol{x}_0\) by
\[ \hat{y}_0 = {\mathrm E}[Y |\hat{\boldsymbol{\beta}}, \boldsymbol{x}_0] = h(\hat{\eta}_0) = h(\hat{\boldsymbol{\beta}}^T \boldsymbol{x}_0).\]
Now we want to construct confidence intervals for \({\mathrm E}[Y |\boldsymbol{\beta}, \boldsymbol{x}_0]\). To do this, we note that (see exercise 10 in Chapter 2):
\[ {\mathrm{Var}}[\hat{\boldsymbol{\beta}}] \stackrel{a}{=}F(\hat{\boldsymbol{\beta}})^{-1}.\]
So,
\[ {\mathrm{Var}}[\hat{\eta}_0] \stackrel{a}{=}\boldsymbol{x}_0^T \;{\mathrm{Var}}[\hat{\boldsymbol{\beta}}] \;\boldsymbol{x}_0 \stackrel{a}{=}\boldsymbol{x}_0^T \;F(\hat{\boldsymbol{\beta}})^{-1} \;\boldsymbol{x}_0 \]
or alternatively,
\[ \text{SE}[\hat{\eta}_0] \stackrel{a}{=}\sqrt{\boldsymbol{x}_0^T \;F(\hat{\boldsymbol{\beta}})^{-1} \;\boldsymbol{x}_0}. \]
Thus, an approximate \((1 - \alpha)\) confidence interval for \({\mathrm E}[Y |\boldsymbol{\beta}, \boldsymbol{x}_0]\) is
\[ CI = \left[ h \left( \hat{\boldsymbol{\beta}}^T \boldsymbol{x}_0 - z_\frac{\alpha}{2} \sqrt{\boldsymbol{x}_0^T \;F(\hat{\boldsymbol{\beta}})^{-1} \;\boldsymbol{x}_0} \right), \; h \left( \hat{\boldsymbol{\beta}}^T \boldsymbol{x}_0 + z_\frac{\alpha}{2} \sqrt{\boldsymbol{x}_0^T \;F(\hat{\boldsymbol{\beta}})^{-1} \;\boldsymbol{x}_0} \right) \right]. \]
Note that in general, this is not symmetric about \(h(\hat{\boldsymbol{\beta}}^T \boldsymbol{x}_0)\).
What about predictive intervals for \(y_0\), by analogy with the linear model case? These are more complicated, as they depend on the response distribution, and we do not consider them here.
Example: Hospital Stay Data
Consider the GLM in Section 2.10.2. Suppose we would like to predict the duration of stay for a new individual with age \(60\), and temperature \(99\). We would call:
## 1
## 13.4064Note that if we do not specify type="response", the function above will return the linear predictor by default:
## 1
## 2.595732In this case, we will need to manually compute the response:
## 1
## 13.4064Now suppose we want to find the 95% confidence interval for the expected mean function at age 60 and temperature 99. We can do this as follows.
# Compute the predicted linear predictor as above
lphat <- predict(hosp.glm, newdata=data.frame(age=60, temp1=99))
# Extract the covariance matrix, which also equals F(betahat)^(-1)
varhat <- summary(hosp.glm)$cov.scaled
# Define new data point with an additional covariate for the intercept
x0 = c(1, 60, 99)
# Compute the width of the interval for the linear predictor
span <- qnorm(0.975) * sqrt( x0 %*% varhat %*% x0)
# Compute the interval for the mean
c(exp(lphat-span), exp(lphat+span))## [1] 8.836973 20.3386013.2 Hypothesis Tests
Suppose we wish to test the values of \(\hat{\boldsymbol{\beta}}\), just as for linear models. We describe below the simple tests and generalisations of these tests to nested models.
3.2.1 Simple Tests
We take as hypotheses \(\mathcal{H}_0: \boldsymbol{\beta} = \boldsymbol{b}\) and \(\mathcal{H}_1: \boldsymbol{\beta} \neq \boldsymbol{b}\). We can test the hypotheses using one of the following two tests. These tests are related but not exactly the same, as illustrated in the figure below.
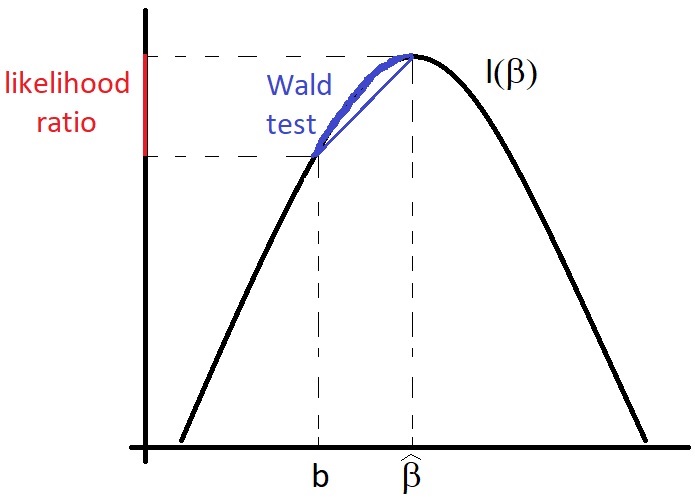
Figure 3.1: Illustration of the relationship between the Wald test and the likelihood ratio test.
3.2.1.1 Wald Test
An obvious candidate for a test statistic is the squared Mahalanobis distance of \(\hat{\boldsymbol{\beta}}\) from \(\boldsymbol{\beta}\), otherwise known as the Wald statistic. Under \(\mathcal{H}_0\), from Equation (2.24) and exercise 10 in Chapter 2, we have:
\[ W = (\hat{\boldsymbol{\beta}}- \boldsymbol{b})^T \;F(\hat{\boldsymbol{\beta}}) \;(\hat{\boldsymbol{\beta}}- \boldsymbol{b}) \stackrel{a}{\sim} \chi^2(p). \tag{3.1} \]
Thus, we reject \(\mathcal{H}_0\) at significance level \(\alpha\) if \(W > \chi^2_{p, \alpha}\).
3.2.1.2 Likelihood Ratio Test
An alternative is a likelihood ratio test. Define
\[ \Lambda = 2\log \left( \frac{L(\hat{\boldsymbol{\beta}})}{L(\boldsymbol{\beta})} \right) = 2(\ell(\hat{\boldsymbol{\beta}}) - \ell(\boldsymbol{\beta})). \]
First, we need to find the distribution of \(\Lambda\). Taylor-expanding \(\ell\), we have:
\[ \ell(\boldsymbol{\beta}) \stackrel{a}{=}\ell(\hat{\boldsymbol{\beta}}) + (\boldsymbol{\beta} - \hat{\boldsymbol{\beta}})^T S(\hat{\boldsymbol{\beta}}) - \frac{1}{2} (\boldsymbol{\beta} - \hat{\boldsymbol{\beta}})^T \;F(\hat{\boldsymbol{\beta}}) \;(\boldsymbol{\beta} - \hat{\boldsymbol{\beta}}). \]
But \(S(\hat{\boldsymbol{\beta}}) = 0\), so we have:
\[ 2(\ell(\hat{\boldsymbol{\beta}}) - \ell(\boldsymbol{\beta})) \stackrel{a}{=}(\boldsymbol{\beta} - \hat{\boldsymbol{\beta}})^T \;F(\hat{\boldsymbol{\beta}}) \;(\boldsymbol{\beta} - \hat{\boldsymbol{\beta}}) \stackrel{a}{\sim} \chi^2(p). \]
Under \(\mathcal{H}_{0}\), we have \(\boldsymbol{\beta} = \boldsymbol{b}\), and thus:
\[ \Lambda = 2(\ell(\hat{\boldsymbol{\beta}}) - \ell(\boldsymbol{b})) \stackrel{a}{\sim} \chi^{2}(p). \tag{3.2} \]
Hence, we reject \(\mathcal{H}_{0}\) at significance level \(\alpha\) if \(\Lambda > \chi^{2}_{p, \alpha}\).
3.2.2 Generalisation to Nested Models
We can generalise the above simple tests to nested models. In particular, our hypotheses are now:
\[\begin{align} \mathcal{H}_0 &: C\boldsymbol{\beta} = \gamma \\ \mathcal{H}_1 &: C\boldsymbol{\beta} \neq \gamma \end{align}\]
where \(C\in{\mathbb R}^{s\times p}\), \(\dim(\text{image}(C)) = s\), and \(\gamma\in{\mathbb R}^{s}\).
Here the equation \(C\boldsymbol{\beta} = \gamma\) constrains the possible values of \(\boldsymbol{\beta}\), reducing the dimensionality of the space of possible solutions by \(s\). The set of \(\boldsymbol{\beta} \in {\mathbb R}^{p}\) satisfying \(C\boldsymbol{\beta} = \gamma\) forms a \((p - s)\)-dimensional affine subspace of \({\mathbb R}^{p}\). Therefore, this corresponds to a restricted or reduced model, as against \(\mathcal{H}_{1}\), which corresponds to the full model. We may sometimes say that \(\mathcal{H}_{0}\) is a submodel of \(\mathcal{H}_{1}\) because the parameter space of \(\mathcal{H}_{0}\) is a subset of the parameter space of \(\mathcal{H}_{1}\).
Example
Let \(\boldsymbol{\beta} \in {\mathbb R}^{p}\) and
\[\begin{align} C & = \begin{pmatrix} 1 & 0 & 0 & \ldots & 0 \\ 0 & 1 & 0 & \ldots & 0 \end{pmatrix} \in {\mathbb R}^{2\times p} \\ \gamma & = 0 \in {\mathbb R}^{2}. \end{align}\]
Then
\[\begin{equation} \mathcal{H}_{0}: \begin{cases} \beta_{1} = 0 & \\ \beta_{2} = 0 & \end{cases} \end{equation}\]
whereas \(\mathcal{H}_{1}\) has \(\beta_{1}\) and \(\beta_{2}\) unrestricted.
For the nested models, we have more general versions of the Wald test and likelihood ratio test below.
3.2.2.1 Wald Test
We have that under \(\mathcal{H}_0\),
\[ W = (C\hat{\boldsymbol{\beta}} - \gamma)^T \; \left( C \;F(\hat{\boldsymbol{\beta}})^{-1} \;C^T \right)^{-1} \; (C\hat{\boldsymbol{\beta}} - \gamma) \stackrel{a}{\sim} \chi^{2}(s) \]
where, recall, \(s\) is the number of constraints; or, equivalently, the difference in the number of parameters; or more abstractly, the difference in the dimensions of the parameter spaces. Thus, we reject \(\mathcal{H}_0\) at significance level \(\alpha\) if \(W > \chi^2_{s, \alpha}\).
3.2.2.2 Likelihood Ratio Test
We have that under \(\mathcal{H}_0\),
\[ \Lambda = 2(\ell(\hat{\boldsymbol{\beta}}) - \ell(\tilde{\boldsymbol{\beta}})) \stackrel{a}{\sim} \chi^{2}(s) \]
where \(\hat{\boldsymbol{\beta}}\) is the MLE under \(\mathcal{H}_{1}\), while \(\tilde{\boldsymbol{\beta}}\) is the MLE under \(\mathcal{H}_{0}\), that is, the MLE for the restricted model. Thus, we reject \(\mathcal{H}_0\) at significance level \(\alpha\) if \(\Lambda > \chi^2_{s, \alpha}\).
Note that compared to the likelihood ratio test, the Wald test does not require the MLE under \(\mathcal{H}_{0}\). Whether this is a good thing is open to question.
3.2.3 Example: Hospital Stay Data
Consider again the hospital stay data. We construct a Gamma GLM with log link for the duration of hospital stay as a function of age and temp1, the temperature at admission, as follows.
\[\begin{equation} \eta = \beta_{1} + \beta_{2}\texttt{age} + \beta_{3}\texttt{temp1} \end{equation}\]
where \[\begin{equation} \texttt{duration} |\texttt{age}, \texttt{temp1} \sim \text{Gamma}(\nu, \nu e^{-\eta}). \end{equation}\]
This model can be coded in R using:
data(hosp, package="npmlreg")
hosp.glm <- glm(duration~age + temp1, data=hosp, family=Gamma(link=log))Note that the chosen parameterisation means that, from properties of the Gamma distribution:
\[\begin{align} \textrm{E}[\texttt{duration} |\texttt{age}, \texttt{temp1}] & = {\nu \over \nu e^{-\eta}} = e^{\eta} \\ \textrm{Var}[\texttt{duration} |\texttt{age}, \texttt{temp1}] & = {\nu \over (\nu e^{-\eta})^{2}} = {e^{2\eta} \over \nu}. \end{align}\]
The first equation says that we are using a log link, i.e. an exponential response; the second equation identifies \(\displaystyle \phi = \frac{1}{\nu}\) and \(\mathcal{V}(\mu) = \mu^{2}\).
We now wish to test \[ \mathcal{H}_{0}: \beta_{3} = 0\] against \[ \mathcal{H}_{1}: \beta_{3} \neq 0. \]
We note that \[ C = \begin{pmatrix} 0 & 0 & 1 \end{pmatrix} \in {\mathbb R}^{1 \times 3} \]
while \(\gamma = 0\), so that the constraint equation can be written as \[ \begin{pmatrix} 0 & 0 & 1 \end{pmatrix} \begin{pmatrix} \beta_{1} \\ \beta_{2} \\ \beta_{3} \end{pmatrix} = 0. \]
The variance we are looking for is
\[ C\;F(\hat{\boldsymbol{\beta}})^{-1} \;C^{T} = \begin{pmatrix} 0 & 0 & 1 \end{pmatrix} F(\hat{\boldsymbol{\beta}})^{-1} \begin{pmatrix} 0 \\ 0 \\ 1 \end{pmatrix} = \text{Var}[\hat{\boldsymbol{\beta}}_{3}] = 0.028 \]
which can be obtained from R using:
## (Intercept) age temp1
## (Intercept) 276.25822713 -3.728838e-02 -2.7943778049
## age -0.03728838 3.246846e-05 0.0003656812
## temp1 -2.79437780 3.656812e-04 0.0282713219The Wald statistic is then given by \[ W = {\hat{\boldsymbol{\beta}}_{3}^{2} \over \textrm{Var}[\hat{\boldsymbol{\beta}}_{3}]} = {(0.31)^{2} \over 0.028} = 3.32. \]
Since \(\chi^{2}_{1, 0.05} = 3.84\) and \(\chi^{2}_{1, 0.1} = 2.71\), we see that we do not reject \(\mathcal{H}_{0}\) at the \(5\%\) level, but do reject at the \(10\%\) level.
3.2.3.1 Does R give what one expects?
Notice that when \(\phi\) has to be estimated together with \(\boldsymbol{\beta}\), we have a situation similar to an unknown variance in the testing of means, going under the title “small sample t-tests.”
For the example above, we have \[ \sqrt{W} = {\hat{\boldsymbol{\beta}}_{3} \over \text{SE}(\hat{\boldsymbol{\beta}}_{3})} = {0.31 \over 0.17} = 1.82. \]
This is the same as the number in the “\(t\)-value” column in the R summary:
##
## Call:
## glm(formula = duration ~ age + temp1, family = Gamma(link = log),
## data = hosp)
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -28.654096 16.621018 -1.724 0.0987 .
## age 0.014900 0.005698 2.615 0.0158 *
## temp1 0.306624 0.168141 1.824 0.0818 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for Gamma family taken to be 0.2690233)
##
## Null deviance: 8.1722 on 24 degrees of freedom
## Residual deviance: 5.7849 on 22 degrees of freedom
## AIC: 142.73
##
## Number of Fisher Scoring iterations: 6However, if \(W \sim \chi^{2}(1)\), then \(\sqrt{W} \sim {\mathcal N}(0, 1)\), leading to \[ p = 2(1 - \Phi(1.82)) = 0.068. \]
This number does not appear in the R summary. The explanation is that if \(\phi\) is estimated, R uses \(t_{n - p}\) rather than \({\mathcal N}(0, 1)\), leading to \[ p = 2(1 - \Phi_{t}(1.82)) = 0.082 \]
and this number does appear in the R summary.
The use of the \(t\) distribution rather than the Gaussian distribution accounts for the extra variability introduced by estimating \(\phi\). It still uses asymptotic normality as a foundation. In fact, R also allows one to assume the dispersion is known:
##
## Call:
## glm(formula = duration ~ age + temp1, family = Gamma(link = log),
## data = hosp)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -28.654096 16.621017 -1.724 0.08471 .
## age 0.014900 0.005698 2.615 0.00892 **
## temp1 0.306624 0.168141 1.824 0.06821 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for Gamma family taken to be 0.2690233)
##
## Null deviance: 8.1722 on 24 degrees of freedom
## Residual deviance: 5.7849 on 22 degrees of freedom
## AIC: 142.73
##
## Number of Fisher Scoring iterations: 6The resulting summary displays a \(z\)-value rather than a \(t\)-value, and computes the corresponding \(p\) using a Gaussian distribution.
We will use \(\chi^{2}\) tests exclusively, thereby ignoring the variability introduced by the estimation of \(\phi\).
3.3 Confidence Regions for \(\hat{\boldsymbol{\beta}}\)
A \((1 - \alpha)\) confidence region for \(\hat{\boldsymbol{\beta}}\) is the set of points in the parameter space \({\mathbb R}^p\) that has probability \((1 - \alpha)\) of containing the true value \(\boldsymbol{\beta}^*\). There are two popular types of confidence regions, which in general are not equivalent.
3.3.1 Hessian Confidence Region
From Equation (3.1), we can construct the \((1 - \alpha)\) Hessian confidence region as: \[ R^{H}_{1 - \alpha} = \left\{\boldsymbol{\beta} :\,(\hat{\boldsymbol{\beta}} - \boldsymbol{\beta})^{T}F(\hat{\boldsymbol{\beta}})(\hat{\boldsymbol{\beta}} - \boldsymbol{\beta}) \leq \chi^{2}_{p,\alpha}\right\}. \]
3.3.2 Method of Support Confidence Region
From Equation (3.2), we can construct the \((1 - \alpha)\) method of support confidence region as: \[ R_{1 - \alpha} = \left\{\boldsymbol{\beta} :\,\ell(\boldsymbol{\beta}) \geq \ell(\hat{\boldsymbol{\beta}}) - \frac{1}{2} \chi^{2}_{p,\alpha}\right\}. \]
3.4 Issues with GLMs and the Wald Test
3.4.1 Separation
Consider a logistic regression problem with linear predictor: \(\eta = \beta_{1} + \beta_{2} x\). Suppose that the data has the following property (not so unreasonable): the \(x\) values of all the points with \(y = 0\) are less than the \(x\) values of all the points with \(y = 1\). This is illustrated in Figure 3.2.
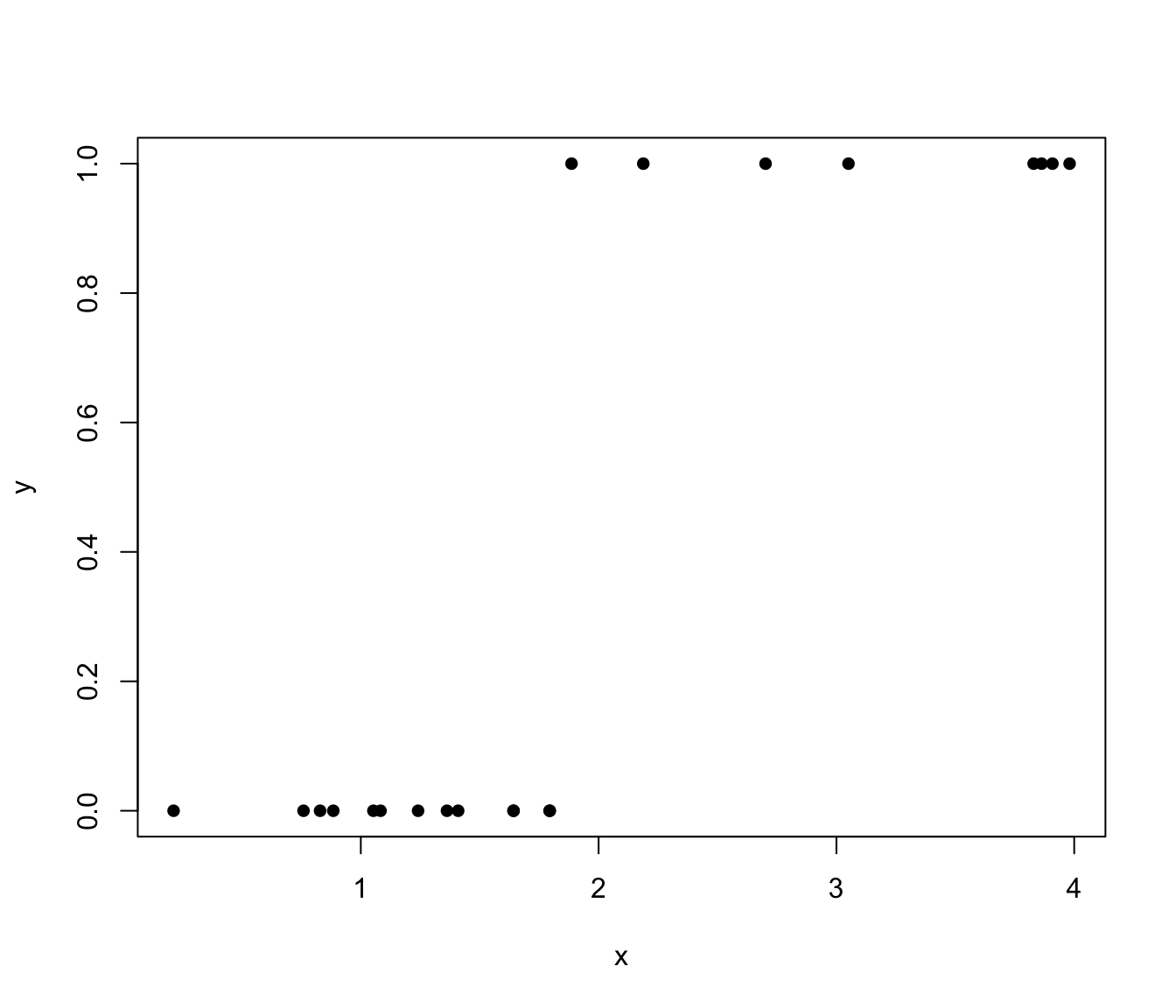
Figure 3.2: Illustration of `separated’ binary data.
Problem
What will be the estimated value \(\hat{\beta}_{2}\) of \(\beta_{2}\)? This question seems unanswerable, but in fact has a very simple answer. First, consider reparameterising the linear predictor by defining
\[\begin{align} \omega & = \beta_{2} \\ x_{0} & = - \frac{\beta_{1}}{\beta_{2}}. \end{align}\]
The linear predictor is thus \[ \eta = \omega (x - x_{0}). \]
The expression for the mean, that is, the probability that \(y = 1\) given \(x\), is then \[ \pi(x) = \frac{e^{\omega (x - x_{0})}}{1 + e^{\omega (x - x_{0})}}. \]
The estimation task is to pick values of \(\omega\) and \(x_{0}\) that maximize the probability of the data. Clearly, if we can choose the parameters so that \(\pi(x) = 1\) for those points with \(y = 1\) and \(\pi(x) = 0\) for those points with \(y = 0\), we cannot do better: this is the maximum achievable with the model, equivalent to the saturated model in fact. We call this a perfect fit.
Now consider the following. Pick \(x_{0}\) so that it lies between the \(x\) with \(y = 0\) and the \(x\) with \(y = 1\). This must be possible because of the initial assumption about the data. Now note that for all the \(x\) with \(y = 0\), \((x - x_{0}) < 0\). If we let \(\omega\rightarrow\infty\), then \(\pi(x)\rightarrow 0\). On the other hand, for all the \(x\) with \(y = 1\), \((x - x_{0}) > 0\), so that as \(\omega\rightarrow\infty\), \(\pi(x)\rightarrow 1\). The limiting solution is thus a step function with the step at \(x_{0}\).
We can therefore achieve a perfect fit by allowing \(\omega\rightarrow\infty\). Unfortunately, this means that the linear predictor is not defined, and, practically speaking, the estimation algorithm will not converge. A secondary problem is also the fact that all values of \(x_{0}\) between the two groups of \(x\) values are equivalent, and there is thus no way to pick the correct one.
Of course, in this simple case, we can see what is happening, and can anticipate that a step function might be a solution. In general, however, this situation might be hard to detect, and hard to correct, at least within the framework of GLMs. So what is to be done?
Solution
Remember that we set out to model functional relationships. GLMs are one way to do this, by constraining the form of the function in a useful way. In this case, however, they seem to be too limiting. There are two reasons why this might be the case.
One is that the step function solution is appropriate for the data and context with which we are dealing. In this case, the main problem is that our set of functions is poorly parameterised, and includes the step function only as a singular limiting case. There is no real solution for this in the context of GLMs, although more general models could be used.
The other is that the step function solution is not appropriate, and that we really would expect a smoother solution. This is much harder to deal with in the context of classical statistics. We are saying that we expect the value of \(\omega\) to be finite, larger values becoming less and less probable, until in the limit, an infinite value is impossible. The only real way to deal with this situation is via a prior probability distribution on \(\omega\) or by imposing some regularising constraint, but those are another story and another course. Within the GLM world, one has to simply be aware of the possibility of separation, and that it may be caused by overly subdividing the data via categorical variables, that is, essentially by overfitting.
3.4.2 Hauck-Donner Effect
A related but independent effect was noted by Hauck and Donner (1976), where the Wald’s statistic decreases to zero as the distance between the parameter estimate and the null value increases.
To see this, consider \[ W = \frac{\hat{\boldsymbol{\beta}}^{2} }{{\mathrm{Var}}[\hat{\boldsymbol{\beta}}]}. \]
If \(\hat{\boldsymbol{\beta}}\rightarrow\infty\) (e.g. in cases of separation), then it is quite likely that \({\mathrm{Var}}[\hat{\boldsymbol{\beta}}]\rightarrow\infty\) also. The result can be that the test statistic becomes very small, and in fact tends to zero! So, as one’s null hypothesis gets more and more wrong, the Wald statistic gets smaller and smaller, and it gets harder to reject the increasingly wrong null hypothesis.
3.5 Exercises
Question 1
Consider the model for Hospital Stay data in Section 3.2.3. Using the printed model summary and covariance matrix, answer the following questions whilst avoiding using R as much as possible. After answering all the questions, use R to check your answers.
- Predict the duration of stay for a 40-year-old patient at temperature 98.
- Construct the 98% confidence interval for the expected duration of stay of this patient.
- Perform the Wald tests to compare the model with only the intercept (\(\mathcal{H}_0\)) against the full model (\(\mathcal{H}_1\)) at the 5% and 1% significance levels.
Question 2
Consider the models for US Polio data in Section 2.9. Perform the Wald tests with appropriate choices of \(\mathcal{H}_0\) and \(\mathcal{H}_1\) to compare the following pairs of models at 5% and 1% significance levels.
polio.glmandpolio1.glm.polio.glmandpolio2.glm.polio.glmandpolio3.glm.polio1.glmandpolio2.glm.polio1.glmandpolio3.glm.polio2.glmandpolio3.glm.
Question 3
We consider again the exponential GLM in exercise 3 of Chapter 2. You can use the solution of that previous exercise to work on this question.
Suppose that we are given the following observations for 17 leukemia patients on the variables \(\texttt{wbc}\) (white blood cell count) and \(\texttt{time}\) (survival time between diagnosis and deaths in weeks).
| wbc | time |
|---|---|
| 3.36 | 65 |
| 2.88 | 156 |
| 3.63 | 100 |
| 3.41 | 134 |
| 3.78 | 16 |
| 4.02 | 108 |
| 4.00 | 121 |
| 4.23 | 4 |
| 3.73 | 39 |
| 3.85 | 143 |
| 3.97 | 56 |
| 4.51 | 26 |
| 4.54 | 22 |
| 5.00 | 1 |
| 5.00 | 1 |
| 4.72 | 5 |
| 5.00 | 65 |
An exponential GLM of the type above is fitted to these data, with linear predictor \(\eta_i=\beta_1+\beta_2{\tt wbc}_i\). Compute the expected Fisher Information matrix and its inverse.
The (edited) summary of the model fitted in R is given below. Use the summary to calculate an estimate of the expected value of \(\texttt{time}\) as well as an approximate \(95\%\) confidence interval for this expected value, when \(\texttt{wbc} = 3\).
The estimate for \(\beta_2\) is \(\hat{\beta_2} = -1.109\). Interpret this value, and suggest and evaluate a test of \(\mathcal{H}_0: \beta_2=0\) at the \(5\%\) level of significance.
Question 4
In the UK Premier League season 2018-2019, each football team had played 21 games by the 10th January 2019. A simple model is proposed that assigns each team a “strength”, \(\beta_j\), where we model the probability of a home team win by: \[ p(j, k) = P(\text{home team } j \text{ beats away team } k) = \frac{e^{\beta_0 + \beta_j - \beta_k}}{1+e^{\beta_0 + \beta_j - \beta_k}}. \] with \(\beta_0\) being a fixed constant (intercept). The table below gives the intercept and strengths when the above model was fitted.
How would you construct a design matrix in order to treat this as a standard GLM? Explain how to interpret the intercept in this setup.
The next match was on 12th January 2019, when Newcastle were defeated playing away against Chelsea. Bookies were offering odds on Chelsea winning of 5:1. Does this simple model agree with these odds?
| Team | \(\beta\) |
|---|---|
| Liverpool | \(1.555\) |
| Manchester City | \(1.009\) |
| Tottenham | \(0.063\) |
| Arsenal | — |
| Chelsea | \(-0.498\) |
| Manchester United | \(-0.711\) |
| Watford | \(-0.947\) |
| Wolves | \(-1.117\) |
| Leicester | \(-1.208\) |
| Everton | \(-1.348\) |
| West Ham | \(-1.401\) |
| Newcastle | \(-1.507\) |
| Brighton | \(-1.602\) |
| Bournemouth | \(-1.752\) |
| Cardiff | \(-1.981\) |
| Crystal Palace | \(-2.032\) |
| Burnley | \(-2.165\) |
| Southampton | \(-2.500\) |
| Fulham | \(-2.595\) |
| Huddersfield | \(-2.958\) |
| Intercept, \(\beta_0\) | \(-0.284\) |