Chapter 27 Calculate Motif Deviations and Footprinting
library(ArchR)27.1 Description
calculate motif deviations and footprinting analysis
27.2 Set env and load arrow project
## Section: set default para
##################################################
addArchRThreads(threads = 16) # setting default number of parallel threads
## Section: load object
##################################################
proj <- loadArchRProject(path = "data/ArchR/ArrowProject/Merged/")27.3 chromVAR
## Section: add bg peak
##################################################
proj <- addBgdPeaks(proj)
## Section: add deviations scores
##################################################
proj <- addDeviationsMatrix(
ArchRProj = proj,
peakAnnotation = "Motif",
force = TRUE,
binarize = TRUE
)
saveArchRProject(ArchRProj = proj, load = FALSE)27.4 Visualize motif deviation scores
## Section: visu check
##################################################
plotVarDev <- getVarDeviations(proj, name = "MotifMatrix", plot = TRUE)## DataFrame with 6 rows and 6 columns
## seqnames idx name combinedVars combinedMeans rank
## <Rle> <array> <array> <numeric> <numeric> <integer>
## f157 z 157 ZEB1_157 7.67441937112557 0.0297632393524911 1
## f97 z 97 TCF4_97 7.32288151518124 0.0801777298018509 2
## f38 z 38 ID3_38 7.15062258662861 0.0707725670149615 3
## f75 z 75 ID4_75 7.15062258662861 0.0707725670149615 4
## f69 z 69 MESP1_69 6.23547776291339 0.0637223302658248 5
## f94 z 94 MESP2_94 6.23547776291339 0.0637223302658248 6motifs <- c("PAX6", "EOMES", "NEUROD2", "DLX2")
markerMotifs <- getFeatures(proj, select = paste(motifs, collapse="|"), useMatrix = "MotifMatrix")
markerMotifs## [1] "z:EOMES_788" "z:PAX6_604" "z:DLX2_438"
## [4] "z:NEUROD2_73" "deviations:EOMES_788" "deviations:PAX6_604"
## [7] "deviations:DLX2_438" "deviations:NEUROD2_73"markerMotifs <- grep("z:", markerMotifs, value = TRUE)
markerMotifs## [1] "z:EOMES_788" "z:PAX6_604" "z:DLX2_438" "z:NEUROD2_73"markerRNA <- getFeatures(proj, select = paste(motifs, collapse="|"), useMatrix = "GeneScoreMatrix")
markerRNA## [1] "DLX2" "PAX6" "NEUROD2" "EOMES"p <- plotEmbedding(
ArchRProj = proj,
colorBy = "MotifMatrix",
name = markerMotifs[c(2,1,4,3)],
embedding = "UMAP_peak",
imputeWeights = getImputeWeights(proj),
pal = ArchRPalettes$solarExtra
)## Getting ImputeWeights## No imputeWeights found, returning NULL## ArchR logging to : ArchRLogs/ArchR-plotEmbedding-2638580bffa0-Date-2021-11-12_Time-15-00-37.log
## If there is an issue, please report to github with logFile!## Getting UMAP Embedding## ColorBy = MotifMatrix## Getting Matrix Values...## 2021-11-12 15:00:40 :## ## Plotting Embedding## 1 2 3 4
## ArchR logging successful to : ArchRLogs/ArchR-plotEmbedding-2638580bffa0-Date-2021-11-12_Time-15-00-37.logp2 <- lapply(p, function(x){
x + guides(color = FALSE, fill = FALSE) +
theme_ArchR(baseSize = 6.5) +
theme(plot.margin = unit(c(0, 0, 0, 0), "cm")) +
theme(
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)
})
p3 <- plotEmbedding(
ArchRProj = proj,
colorBy = "GeneScoreMatrix",
name = markerRNA[c(2,4,3,1)],
embedding = "UMAP_peak",
imputeWeights = getImputeWeights(proj),
pal = ArchRPalettes$coolwarm
)## Getting ImputeWeights
## No imputeWeights found, returning NULL
## ArchR logging to : ArchRLogs/ArchR-plotEmbedding-26386c03ed6e-Date-2021-11-12_Time-15-00-45.log
## If there is an issue, please report to github with logFile!
## Getting UMAP Embedding
## ColorBy = GeneScoreMatrix
## Getting Matrix Values...
## 2021-11-12 15:00:45 :
##
## Plotting Embedding
## 1 2 3 4
## ArchR logging successful to : ArchRLogs/ArchR-plotEmbedding-26386c03ed6e-Date-2021-11-12_Time-15-00-45.logp4 <- lapply(p3, function(x){
x + guides(color = FALSE, fill = FALSE) +
theme_ArchR(baseSize = 6.5) +
theme(plot.margin = unit(c(0, 0, 0, 0), "cm")) +
theme(
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)
})
do.call(cowplot::plot_grid, c(list(ncol = 4),p2))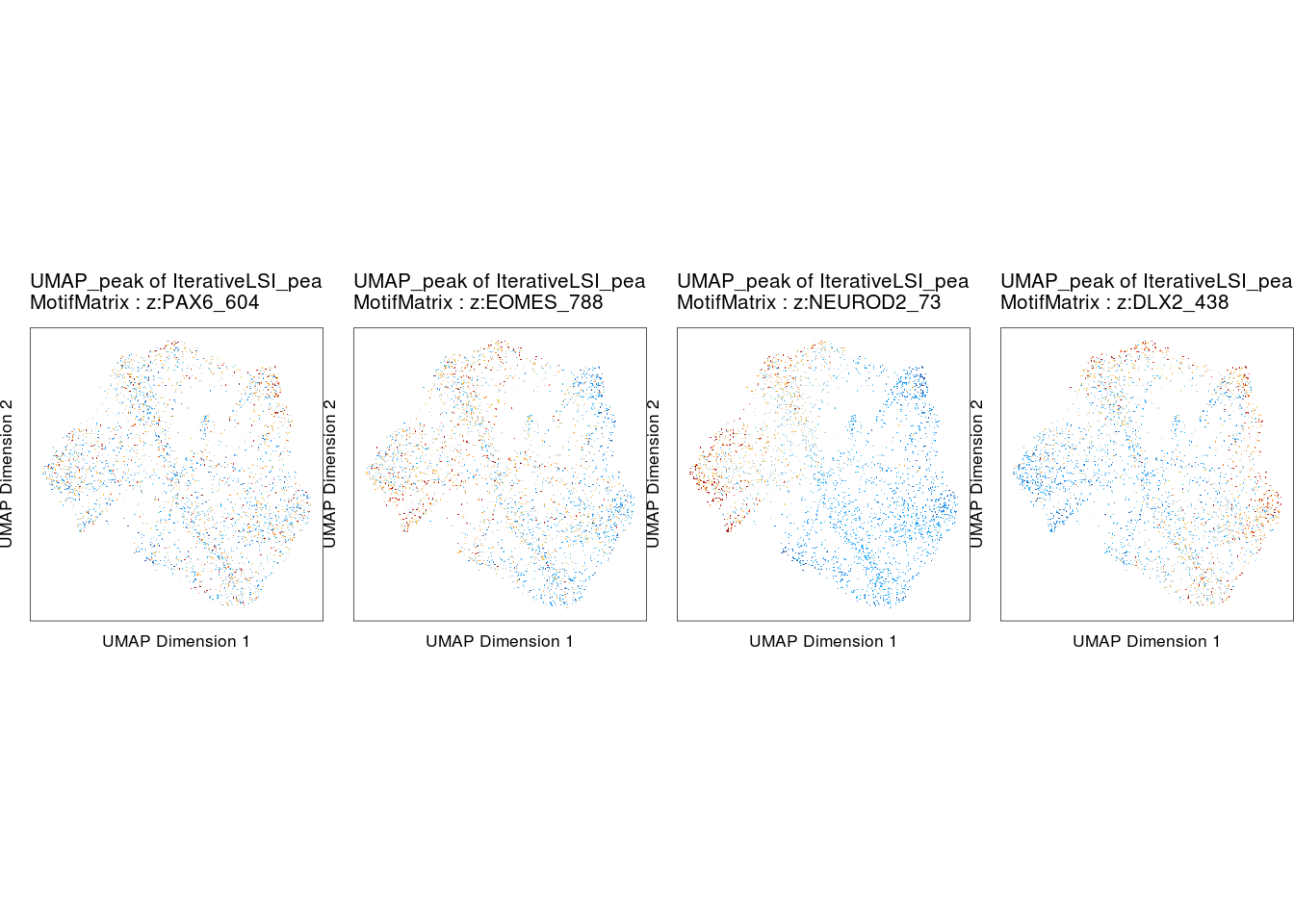
do.call(cowplot::plot_grid, c(list(ncol = 4),p4))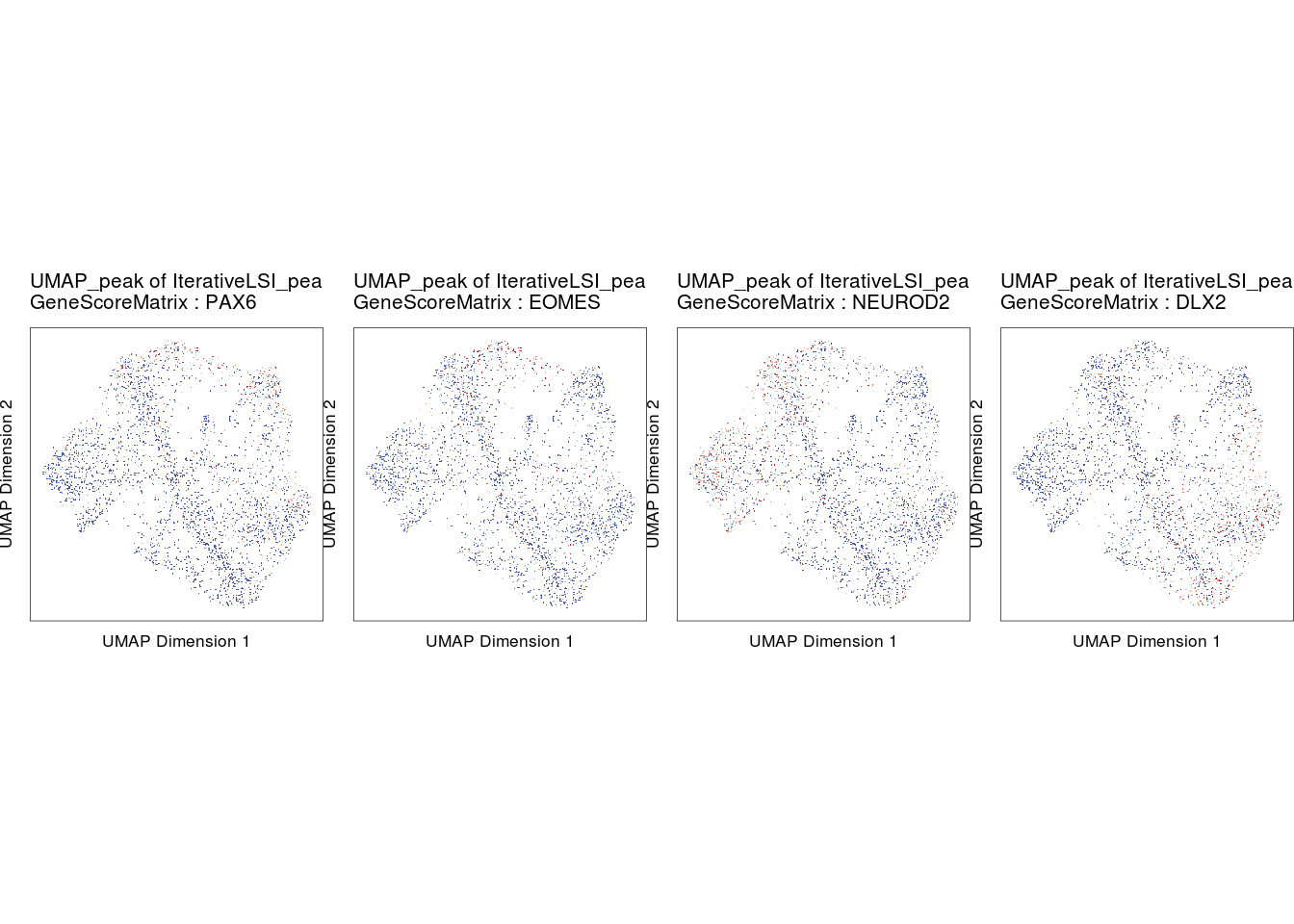
27.5 Footprint
## Section: foot print
##################################################
motifPositions <- getPositions(proj)
motifPositions## GRangesList object of length 870:
## $TFAP2B_1
## GRanges object with 28983 ranges and 1 metadata column:
## seqnames ranges strand | score
## <Rle> <IRanges> <Rle> | <numeric>
## [1] chr1 71450-71461 - | 8.15862455767063
## [2] chr1 113748-113759 + | 8.07432085875437
## [3] chr1 267338-267349 + | 9.77203962986206
## [4] chr1 267338-267349 - | 8.26644472169785
## [5] chr1 1642359-1642370 - | 8.3241486956364
## ... ... ... ... . ...
## [28979] chrX 150696503-150696514 + | 9.08449223764575
## [28980] chrX 150696503-150696514 - | 9.23118846536946
## [28981] chrX 150796823-150796834 + | 8.21870363758565
## [28982] chrX 151250912-151250923 + | 8.43788710872478
## [28983] chrX 151560217-151560228 + | 8.06918260206019
## -------
## seqinfo: 21 sequences from an unspecified genome; no seqlengths
##
## ...
## <869 more elements>motifs <- c("NFIX", "EOMES", "NEUROD2", "DLX2")
markerMotifs <- unlist(lapply(motifs, function(x) grep(x, names(motifPositions), value = TRUE)))
markerMotifs## [1] "NFIX_738" "EOMES_788" "NEUROD2_73" "DLX2_438"#proj <- addGroupCoverages(ArchRProj = proj, groupBy = "BioCluster")
seFoot <- getFootprints(
ArchRProj = proj,
positions = motifPositions[markerMotifs],
groupBy = "predictedGroup_Un"
)## ArchR logging to : ArchRLogs/ArchR-getFootprints-263874055a4e-Date-2021-11-12_Time-15-00-56.log
## If there is an issue, please report to github with logFile!## 2021-11-12 15:01:02 : Computing Kmer Bias Table, 0.101 mins elapsed.## 2021-11-12 15:02:31 : Finished Computing Kmer Tables, 1.492 mins elapsed.## 2021-11-12 15:02:31 : Computing Footprints, 1.592 mins elapsed.## 2021-11-12 15:02:40 : Computing Footprints Bias, 1.749 mins elapsed.## 2021-11-12 15:02:48 : Summarizing Footprints, 1.871 mins elapsed.plotFootprints(
seFoot = seFoot,
ArchRProj = proj,
normMethod = "Subtract",
plotName = "Footprints-Subtract-Bias",
addDOC = FALSE,
smoothWindow = 5
)## ArchR logging to : ArchRLogs/ArchR-plotFootprints-2638137b252-Date-2021-11-12_Time-15-02-50.log
## If there is an issue, please report to github with logFile!## 2021-11-12 15:02:51 : Plotting Footprint : NFIX_738 (1 of 4), 0.005 mins elapsed.## Applying smoothing window to footprint## Normalizing by flanking regions## NormMethod = Subtract## 2021-11-12 15:02:56 : Plotting Footprint : EOMES_788 (2 of 4), 0.089 mins elapsed.## Applying smoothing window to footprint## Normalizing by flanking regions## NormMethod = Subtract## 2021-11-12 15:03:01 : Plotting Footprint : NEUROD2_73 (3 of 4), 0.171 mins elapsed.## Applying smoothing window to footprint## Normalizing by flanking regions## NormMethod = Subtract## 2021-11-12 15:03:04 : Plotting Footprint : DLX2_438 (4 of 4), 0.229 mins elapsed.## Applying smoothing window to footprint## Normalizing by flanking regions## NormMethod = Subtract## ArchR logging successful to : ArchRLogs/ArchR-plotFootprints-2638137b252-Date-2021-11-12_Time-15-02-50.log