6 Vraag6: Hox genes
6.1 A. Verkrijg de sequentie van de HOXA1 gen van de mens, bonobo aap en gorilla met behulp van Biomart
#Mart van vraag 1 nog beschikbaar:
#h_sapiens
#finding out names of marts for gorilla and bonobo
searchDatasets(h_sapiens, pattern = "(B|b)onobo")## dataset description version
## 154 ppaniscus_gene_ensembl Bonobo genes (panpan1.1) panpan1.1searchDatasets(h_sapiens, pattern = "(G|g)orilla")## dataset description version
## 74 ggorilla_gene_ensembl Gorilla genes (gorGor4) gorGor4#getting marts
p_paniscus <- useMart(biomart = "ensembl", dataset = "ppaniscus_gene_ensembl")
g_gorilla <- useMart(biomart = "ensembl", dataset = "ggorilla_gene_ensembl")
#getting necessary filters and attributes
searchFilters(h_sapiens, pattern = "(G|g)ene.*name")## name description
## 18 with_entrezgene_trans_name With EntrezGene transcript name ID(s)
## 64 external_gene_name Gene Name(s) [e.g. MT-TF]
## 76 entrezgene_trans_name EntrezGene transcript name ID(s) [e.g. AA06-201]
## 119 wikigene_name WikiGene name(s) [e.g. A1BG]searchAttributes(h_sapiens, pattern = "ppaniscus.*homolog")## name
## 626 ppaniscus_homolog_ensembl_gene
## 627 ppaniscus_homolog_associated_gene_name
## 628 ppaniscus_homolog_ensembl_peptide
## 629 ppaniscus_homolog_chromosome
## 630 ppaniscus_homolog_chrom_start
## 631 ppaniscus_homolog_chrom_end
## 632 ppaniscus_homolog_canonical_transcript_protein
## 633 ppaniscus_homolog_subtype
## 634 ppaniscus_homolog_orthology_type
## 635 ppaniscus_homolog_perc_id
## 636 ppaniscus_homolog_perc_id_r1
## 637 ppaniscus_homolog_goc_score
## 638 ppaniscus_homolog_wga_coverage
## 639 ppaniscus_homolog_orthology_confidence
## description page
## 626 Bonobo gene stable ID homologs
## 627 Bonobo gene name homologs
## 628 Bonobo protein or transcript stable ID homologs
## 629 Bonobo chromosome/scaffold name homologs
## 630 Bonobo chromosome/scaffold start (bp) homologs
## 631 Bonobo chromosome/scaffold end (bp) homologs
## 632 Query protein or transcript ID homologs
## 633 Last common ancestor with Bonobo homologs
## 634 Bonobo homology type homologs
## 635 %id. target Bonobo gene identical to query gene homologs
## 636 %id. query gene identical to target Bonobo gene homologs
## 637 Bonobo Gene-order conservation score homologs
## 638 Bonobo Whole-genome alignment coverage homologs
## 639 Bonobo orthology confidence [0 low, 1 high] homologssearchAttributes(h_sapiens, pattern = "ggorilla.*homolog")## name
## 1428 ggorilla_homolog_ensembl_gene
## 1429 ggorilla_homolog_associated_gene_name
## 1430 ggorilla_homolog_ensembl_peptide
## 1431 ggorilla_homolog_chromosome
## 1432 ggorilla_homolog_chrom_start
## 1433 ggorilla_homolog_chrom_end
## 1434 ggorilla_homolog_canonical_transcript_protein
## 1435 ggorilla_homolog_subtype
## 1436 ggorilla_homolog_orthology_type
## 1437 ggorilla_homolog_perc_id
## 1438 ggorilla_homolog_perc_id_r1
## 1439 ggorilla_homolog_goc_score
## 1440 ggorilla_homolog_wga_coverage
## 1441 ggorilla_homolog_orthology_confidence
## description page
## 1428 Gorilla gene stable ID homologs
## 1429 Gorilla gene name homologs
## 1430 Gorilla protein or transcript stable ID homologs
## 1431 Gorilla chromosome/scaffold name homologs
## 1432 Gorilla chromosome/scaffold start (bp) homologs
## 1433 Gorilla chromosome/scaffold end (bp) homologs
## 1434 Query protein or transcript ID homologs
## 1435 Last common ancestor with Gorilla homologs
## 1436 Gorilla homology type homologs
## 1437 %id. target Gorilla gene identical to query gene homologs
## 1438 %id. query gene identical to target Gorilla gene homologs
## 1439 Gorilla Gene-order conservation score homologs
## 1440 Gorilla Whole-genome alignment coverage homologs
## 1441 Gorilla orthology confidence [0 low, 1 high] homologs#attributes: ggorilla_homolog_ensembl_gene, ppaniscus_homolog_ensembl_gene, external_gene_id
#filter: external_gene_name
gene_id <- getBM(attributes = c("ensembl_gene_id", "ggorilla_homolog_ensembl_gene", "ppaniscus_homolog_ensembl_gene"),
filters = "external_gene_name",
values = "HOXA1",
mart = h_sapiens)
gene_id %>% kable()| ensembl_gene_id | ggorilla_homolog_ensembl_gene | ppaniscus_homolog_ensembl_gene |
|---|---|---|
| ENSG00000105991 | ENSGGOG00000002209 | ENSPPAG00000000155 |
h_sapiens_HOXA1 <- biomaRt::getSequence(id = "ENSG00000105991", type = "ensembl_gene_id", seqType = "coding", mart = h_sapiens)
g_gorilla_HOXA1 <- biomaRt::getSequence(id = "ENSGGOG00000002209", type = "ensembl_gene_id", seqType = "coding", mart = g_gorilla)
p_paniscus_HOXA1 <- biomaRt::getSequence(id = "ENSPPAG00000000155", type = "ensembl_gene_id", seqType = "coding", mart = p_paniscus)
#checking for duplicates
for (i in c("h_sapiens_HOXA1", "g_gorilla_HOXA1", "p_paniscus_HOXA1")){
print(c(i, duplicated(as.vector(get(i)$coding))))
}## [1] "h_sapiens_HOXA1" "FALSE" "FALSE"
## [1] "g_gorilla_HOXA1" "FALSE" "FALSE"
## [1] "p_paniscus_HOXA1" "FALSE" "FALSE"The sequences have been retrieved from the ensembl database, but per ensembl gene id there are two unique coding sequences
table_coding <- bind_rows(h_sapiens_HOXA1, g_gorilla_HOXA1, p_paniscus_HOXA1)
table_coding <- bind_cols(table_coding, tibble(species = c("h_sapiens",
"h_sapiens",
"g_gorilla",
"g_gorilla",
"p_paniscus",
"p_paniscus")))
table_coding %>%
knitr::kable() %>%
column_spec(column = 1, width = "10cm")| coding | ensembl_gene_id | species |
|---|---|---|
| ATGGACAATGCAAGAATGAACTCCTTCCTGGAATACCCCATACTTAGCAGTGGCGACTCGGGGACCTGCTCAGCCCGAGCCTACCCCTCGGACCATAGGATTACAACTTTCCAGTCGTGCGCGGTCAGCGCCAACAGTTGCGGCGGCGACGACCGCTTCCTAGTGGGCAGGGGGGTGCAGATCGGTTCGCCCCACCACCACCACCACCACCACCATCGCCACCCCCAGCCGGCTACCTACCAGACTTCCGGGAACCTGGGGGTGTCCTACTCCCACTCAAGTTGTGGTCCAAGCTATGGCTCACAGAACTTCAGTGCGCCTTACAGCCCCTACGCGTTAAATCAGGAAGCAGACCCACCAAGAAGCCTGTCGCTCCCCCGCATCGGAGACATCTTCTCCAGCGCAGACTTTTGA | ENSG00000105991 | h_sapiens |
| ATGGACAATGCAAGAATGAACTCCTTCCTGGAATACCCCATACTTAGCAGTGGCGACTCGGGGACCTGCTCAGCCCGAGCCTACCCCTCGGACCATAGGATTACAACTTTCCAGTCGTGCGCGGTCAGCGCCAACAGTTGCGGCGGCGACGACCGCTTCCTAGTGGGCAGGGGGGTGCAGATCGGTTCGCCCCACCACCACCACCACCACCACCATCGCCACCCCCAGCCGGCTACCTACCAGACTTCCGGGAACCTGGGGGTGTCCTACTCCCACTCAAGTTGTGGTCCAAGCTATGGCTCACAGAACTTCAGTGCGCCTTACAGCCCCTACGCGTTAAATCAGGAAGCAGACGTAAGTGGTGGGTACCCCCAGTGCGCTCCCGCTGTTTACTCTGGAAATCTCTCATCTCCCATGGTCCAGCATCACCACCACCACCAGGGTTATGCTGGGGGCGCGGTGGGCTCGCCTCAATACATTCACCACTCATATGGACAGGAGCACCAGAGCCTGGCCCTGGCTACGTATAATAACTCCTTGTCCCCTCTCCACGCCAGCCACCAAGAAGCCTGTCGCTCCCCCGCATCGGAGACATCTTCTCCAGCGCAGACTTTTGACTGGATGAAAGTCAAAAGAAACCCTCCCAAAACAGGGAAAGTTGGAGAGTACGGCTACCTGGGTCAACCCAACGCGGTGCGCACCAACTTCACTACCAAGCAGCTCACGGAACTGGAGAAGGAGTTCCACTTCAACAAGTACCTGACGCGCGCCCGCAGGGTGGAGATCGCTGCATCCCTGCAGCTCAACGAGACCCAAGTGAAGATCTGGTTCCAGAACCGCCGAATGAAGCAAAAGAAACGTGAGAAGGAGGGTCTCTTGCCCATCTCTCCGGCCACCCCGCCAGGAAACGACGAGAAGGCCGAGGAATCCTCAGAGAAGTCCAGCTCTTCGCCCTGCGTTCCTTCCCCGGGGTCTTCTACCTCAGACACTCTGACTACCTCCCACTGA | ENSG00000105991 | h_sapiens |
| ATGGACAATGCAAGAATGAACTCCTTCCTGGAATACCCTATACTTAGCAGTGGCGACTCGGGGACCTGCTCAGCCCGAGCCTACCCCTCGGACCATGGGATTACAACTTTCCAGTCGTGCGCGGTCAGTGCCAACAGTTGCGGCGGCGACGACCGCTTCCTAGTGGGCAGGGGGGTGCAGATTGGTTCGCCCCACCAGCACCACCACCACCACCATCACCACCCCCAGCCGGCTACCTACCAGACTTCCGGGAACCTGGGGGTGTCCTACTCCCACTCGAGTTGTGGTCCAAGCTATGGCTCACAGAACTTCAGTGCGCCTTACAGCCCCTACGCGTTAAATCAGGAAGCAGACGTAAGTGGTGGGTACCCCCAGTGCGCTCCCGCTGTTTACTCTGGAAATCTCTCATCTCCCATGGTCCAGCATCACCACCACCACCAGGGTTATGCTGGGGGCGCGGTGGGCTCGCCTCAATACATTCACCACTCATATGGACAGGAGCACCAGAGCCTGGCCCTGGCTACGTATAATAACTCCTTGTCCCCTCTCCACGCCAGCCACCAAGAAGCCTGTCGCTCCCCTGCATCGGAGACATCTTCTCCAGCGCAGACTTTTGACTGGATGAAAGTCAAAAGAAACCCTCCCAAAACAGGGAAAGTTGGAGAGTACGGCTACCTGGGTCAACCCAACGCGGTGCGTACCAACTTCACTACCAAGCAGCTCACGGAACTGGAGAAGGAGTTCCACTTCAACAAGTACCTGACGCGCGCCCGCAGAGTGGAGATCGCTGCATCCCTGCAGCTCAACGAGACCCAAGTGAAGATCTGGTTCCAGAACCGCCGAATGAAGCAAAAGAAACGTGAGAAGGAGGGTCTCTTGCCCATCTCTCCGGCCACCCCGCCAGGAAACGACGAGAAGGCCGAGGAATCCTCAGAGAAGTCCAGCTCTTCGCCCTGCGTTCCTTCCCCGGGGTCTTCTACCTCAGACACTCTGACTACCTCCCACTGA | ENSGGOG00000002209 | g_gorilla |
| ATGGACAATGCAAGAATGAACTCCTTCCTGGAATACCCTATACTTAGCAGTGGCGACTCGGGGACCTGCTCAGCCCGAGCCTACCCCTCGGACCATGGGATTACAACTTTCCAGTCGTGCGCGGTCAGTGCCAACAGTTGCGGCGGCGACGACCGCTTCCTAGTGGGCAGGGGGGTGCAGATTGGTTCGCCCCACCAGCACCACCACCACCACCATCACCACCCCCAGCCGGCTACCTACCAGACTTCCGGGAACCTGGGGGTGTCCTACTCCCACTCGAGTTGTGGTCCAAGCTATGGCTCACAGAACTTCAGTGCGCCTTACAGCCCCTACGCGTTAAATCAGGAAGCAGACCCACCAAGAAGCCTGTCGCTCCCCTGCATCGGAGACATCTTCTCCAGCGCAGACTTTTGA | ENSGGOG00000002209 | g_gorilla |
| ATGGACAATGCAAGAATGAACTCCTTCCTGGAATACCCCATACTTAGCAGTGGCGACTCGGGGACCTGCCCAGCCCGAGCCTACCCCTCGGACCATGGGATTACAACTTTCCAGTCGTGCGCGGTCAGTGCCAACAGTTGCGGCGGCGACGACCGCTTCCTAATGGGCAGGGGGGTGCAGATCGGTTCGCCCCACCACCACCACCACCACCACCACCATCACCACCCCCAGCCGGCTACCTACCAGACTTCCGGGAACCTGGGGGTGTCCTACTCCCACTCGAGTTGTGGTCCAAGCTATGGCTCACAGAACTTCAGTGCGCCTTACAGCCCCTACGCGTTAAATCAGGAAGCAGACGTAAGTGGTGGGTACCCCCAGTGCGCTCCCGCTGTTTACTCTGGAAATCTCTCATCTCCCATGGTCCAGCATCACCACCACCACCAGGGTTATGCTGGGGGCGCGGTGGGCTCGCCTCAATACATTCACCACTCATATGGACAGGAGCACCAGAGCCTGGCCCTGGCTACGTATAATAACTCCTTGTCCCCTCTCCACGCCAGCCACCAAGAAGCCTGTCGCTCCCCTGCATCGGAGACATCTTCTCCAGCGCAGACTTTTGACTGGATGAAAGTCAAAAGAAACCCTCCCAAAACAGGGAAAGTTGGAGAGTACGGCTACCTGGGTCAACCCAACGCGGTGCGCACCAACTTCACTACCAAGCAGCTCACGGAACTGGAGAAGGAGTTCCACTTCAACAAGTACCTGACGCGCGCCCGCAGGGTGGAGATCGCTGCATCCCTGCAGCTCAACGAGACCCAAGTGAAGATCTGGTTCCAGAACCGCCGAATGAAGCAAAAGAAACGTGAGAAGGAGGGTCTCTTGCCCATCTCTCCGGCCACCCCGCCAGGAAACGACGAGAAGGCCGAGGAATCCTCAGAGAAGTCCAGCTCTTCGCCCTGCGTTCCTTCCCCGGGGTCTTCTACCTCAGACACTCTGACTACCTCCCACTGA | ENSPPAG00000000155 | p_paniscus |
| ATGGACAATGCAAGAATGAACTCCTTCCTGGAATACCCCATACTTAGCAGTGGCGACTCGGGGACCTGCCCAGCCCGAGCCTACCCCTCGGACCATGGGATTACAACTTTCCAGTCGTGCGCGGTCAGTGCCAACAGTTGCGGCGGCGACGACCGCTTCCTAATGGGCAGGGGGGTGCAGATCGGTTCGCCCCACCACCACCACCACCACCACCACCATCACCACCCCCAGCCGGCTACCTACCAGACTTCCGGGAACCTGGGGGTGTCCTACTCCCACTCGAGTTGTGGTCCAAGCTATGGCTCACAGAACTTCAGTGCGCCTTACAGCCCCTACGCGTTAAATCAGGAAGCAGACCCACCAAGAAGCCTGTCGCTCCCCTGCATCGGAGACATCTTCTCCAGCGCAGACTTTTGA | ENSPPAG00000000155 | p_paniscus |
6.2 B. Voer een sequence alignment uit op de eiwit sequentie van deze genen
seq <- Biostrings::DNAStringSet(table_coding$coding)
names(seq) <- paste(table_coding$species , table_coding$ensembl_gene_id, sep = "_")
AA <- AlignTranslation(seq, type = "AAStringSet")## Determining distance matrix based on shared 5-mers:
##
|
| | 0%
|
|======================= | 33%
|
|=========================================== | 60%
|
|========================================================= | 80%
|
|================================================================== | 93%
|
|=======================================================================| 100%
##
## Time difference of 0 secs
##
## Clustering into groups by similarity:
##
|
| | 0%
|
|======================= | 33%
|
|=========================================== | 60%
|
|========================================================= | 80%
|
|================================================================== | 93%
|
|=======================================================================| 100%
##
## Time difference of 0 secs
##
## Aligning Sequences:
##
|
| | 0%
|
|============== | 20%
|
|============================ | 40%
|
|=========================================== | 60%
|
|========================================================= | 80%
|
|=======================================================================| 100%
##
## Time difference of 0.03 secs
##
## Iteration 1 of 2:
##
## Determining distance matrix based on alignment:
##
|
| | 0%
|
|======================= | 33%
|
|=========================================== | 60%
|
|========================================================= | 80%
|
|================================================================== | 93%
|
|=======================================================================| 100%
##
## Time difference of 0 secs
##
## Reclustering into groups by similarity:
##
|
| | 0%
|
|======================= | 33%
|
|=========================================== | 60%
|
|========================================================= | 80%
|
|================================================================== | 93%
|
|=======================================================================| 100%
##
## Time difference of 0 secs
##
## Realigning Sequences:
##
|
| | 0%
|
|=======================================================================| 100%
##
## Time difference of 0 secs
##
## Alignment converged - skipping remaining iteration.BrowseSeqs(AA)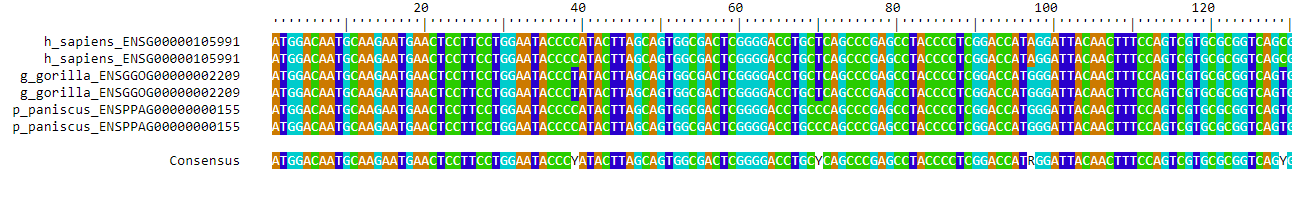
Sequence aligment
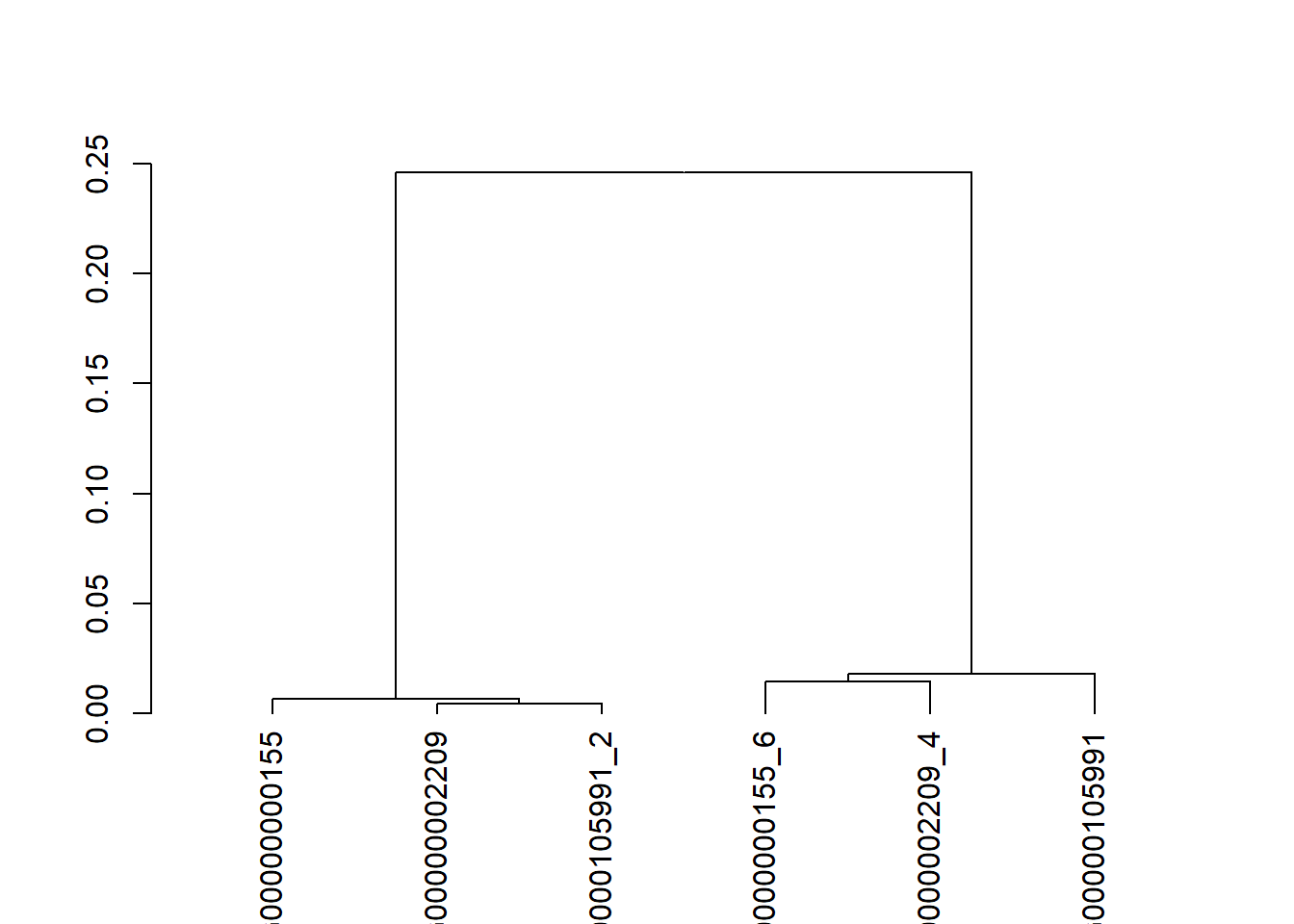
6.3 C. Maak een dendrogram van de eiwit sequenties
#for StaggerAlignment sequences
AA_stag <- StaggerAlignment(AA)## Calculating distance matrix:
##
|
| | 0%
|
|======================= | 33%
|
|=========================================== | 60%
|
|========================================================= | 80%
|
|================================================================== | 93%
|
|=======================================================================| 100%
##
## Time difference of 0 secs
##
## Constructing neighbor-joining tree:
##
|
| | 0%
|
|======================= | 33%
|
|=========================================== | 60%
|
|========================================================= | 80%
|
|================================================================== | 93%
|
|=======================================================================| 100%
##
## Time difference of 0 secs
##
## Staggering insertions and deletions:
##
|
| | 0%
|
|===================== | 30%
|
|========================== | 36%
|
|============================================= | 64%
|
|========================================================= | 80%
|
|========================================================== | 81%
|
|=======================================================================| 100%
##
## Time difference of 0.01 secsAA_dis <- DistanceMatrix(AA_stag)##
|
| | 0%
|
|======================= | 33%
|
|=========================================== | 60%
|
|========================================================= | 80%
|
|================================================================== | 93%
|
|=======================================================================| 100%
##
## Time difference of 0 secsdendogram <- IdClusters(AA_dis, type = "both")##
|
| | 0%
|
|======================= | 33%
|
|=========================================== | 60%
|
|========================================================= | 80%
|
|================================================================== | 93%
|
|=======================================================================| 100%## Warning in IdClusters(AA_dis, type = "both"): Duplicated labels in dendrogram
## appended with index.## Warning in IdClusters(AA_dis, type = "both"): Duplicated labels in myDistMatrix
## appended with index.##
## Time difference of 0 secsplot(dendogram[[2]])