Chapter 5 Electron Transport Chain and Oxidative Phosphorylation
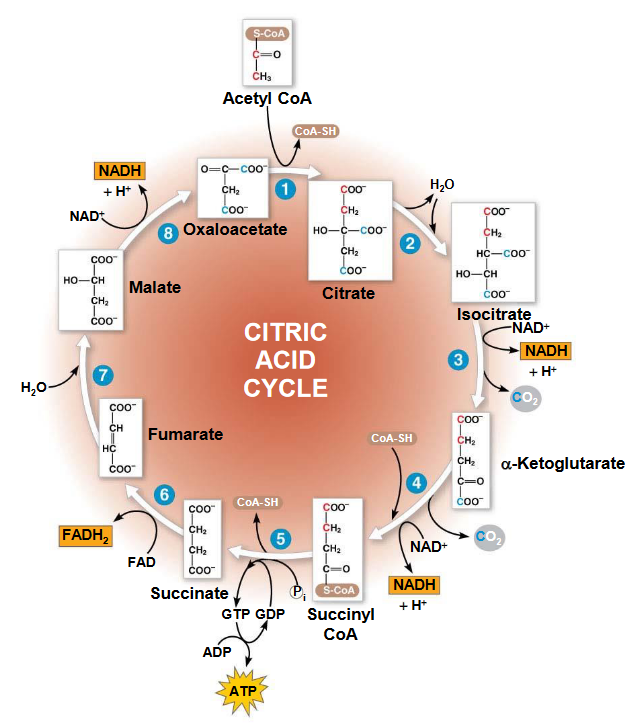
Figure 5.1: Krebs’ Cycle
The ATP yield per molecule of glucose at each stage of cellular respiration is:
- 2 molecules of ATP from glycolysis
- 2 molecules of ATP from the Krebs’ cycle
- 26 - 28 molecules of ATP from the electron transport chain.
Figure 5.2: ATP Yield from Oxidative Phosphorylation
5.1 Some Cursory Knowledge…
5.1.1 Oxidation of glucose
The complete oxidation of one mole of glucose is given by the following reaction:
\[\begin{equation*} C_6H_{12}O_6 + 6O_2 \leftrightarrow 6CO_2 + 6H_2O \text{ | } \Delta G^\omicron = \text{-2823 kJ / mol} \end{equation*}\]
The first half of the above reaction involves the oxidation of carbon atoms:
\[\begin{equation*} C_6H_{12}O_6 + 6O_2 \to 6CO_2 + 24H^+ + 24e^- \end{equation*}\]
The second half, on the contrary, reduces molecular oxygen:
\[\begin{equation*} 6O_2 + 24H^+ + 24e^- \to 12H_2O \end{equation*}\]
Note that the 12 electron pairs (i.e., the e-) in the carbon oxidation portion of the reaction are not transferred to O2 directly, but to the electron carriers NAD+ and FAD to form 2 molecules of FADH2 and 10 molecules of NADH (to participate in the electron transport chain).
5.1.2 Anatomy of the mitochondria
Observe the anatomy of the mitochondria:
Figure 5.3: Mitochondria Anatomy
The inner membrane of the mitochondria is folded into ridges called cristae. Heart muscle cells have a higher amount of mitochondria due to their activity: a huge contrast to liver cells (which have a lesser amounts of mitochondria).
In the picture on the right, the protein complexes I, III, and IV are responsible for pumping protons into the intermembrane space, thereby generating an electrochemical gradient.
5.1.3 Shuttles
5.1.3.1 The malate-aspartate shuttle
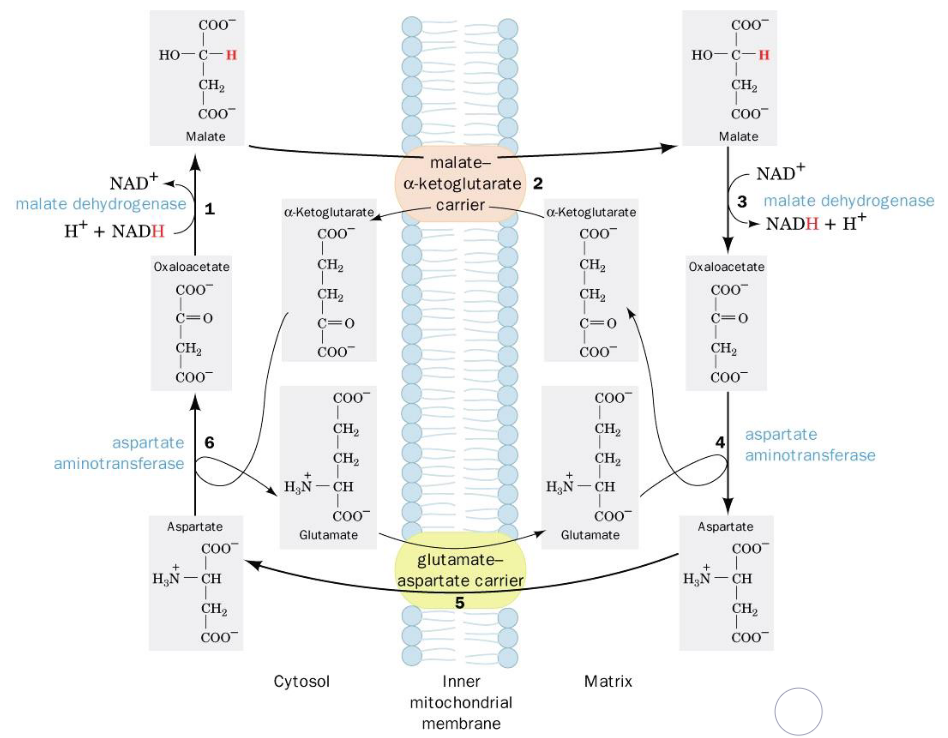
Figure 5.4: Malate-Aspartate Shuttle
Electrons from cytosolic NADH are transported to mitochondrial NADH in steps 1 to 3. Steps 4 to 6 regenerate cytosolic oxalatoacetate.
5.1.3.2 The glycerophosphate shuttle
Figure 5.5: Glycerophosphate Shuttle
Electrons in cytosolic NADH can also be transported to the mitochondria via this shuttle too!
Oxidation of cytosolic NADH by DHAP (i.e., dihydroxyacetone phosphate) is catalyzed by the enzyme glyceraldehyde-3-phosphate. The enzyme flavoprotein dehydrogenase then reduces FAD to FADH2; the latter of which then proceeds to the electron transport chain.
5.1.3.3 The ATP-ADP translocase
Shown on the left is the secondary structure of bovine ATP-ADP translocase. As suggested on the right, the outward transport of ATP is determined by the membrane electrochemical potential:
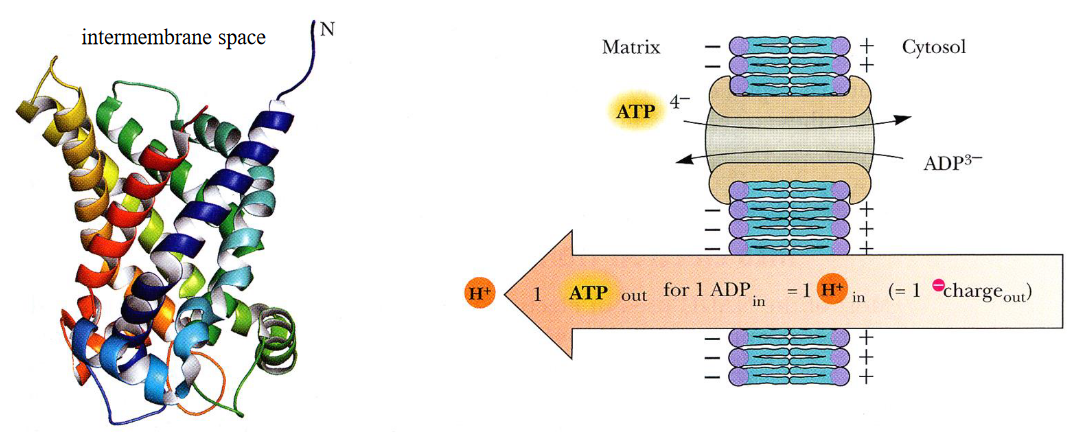
Figure 5.6: ATP-ADP Translocase
This protein accounts for 14% of all mitochondrial proteins. ATP / ADP transport occurs at a single-nucleotide binding site; ATP binds on the matrix side of the protein, causing the protein to reorient itself towards the cytoplasm and exchange ADP for ATP in the end.
ATP carries a -4 charge whereas ADP carries a -3 charge. Hence, we say that there is a net movement of one charge from the matrix to the cytosol.
5.2 The Electron Transport Chain and Redox Potentials
In general, the electron transport chain looks like this:
Figure 5.7: Electron Transport Chain
The electron transport chain is located on the inner membrane of the mitochondria (i.e., the cristae). As seen above, many of the chain’s components are proteins that form multiprotein complexes. Electron carriers in the electron transport chain also alternate between reduced and oxidized states:
Figure 5.8: Proteins in the Electron Transport Chain
When electrons move through the chain, they drop in free energy and are passed through a number of proteins (including cytochromes which have an iron atom bound to them). The chain also does not generate ATP directly (unlike the Krebs’ cycle or glycolysis).
Rather, the chain serves to break the large free-energy drop from food into smaller steps that release the energy in smaller, more appropriate amounts.
5.2.1 How do we measure redox potentials?
A redox reaction can be re-written into two half-equations, both of which are represented as reduction equations:
\[\begin{equation*} A^{n+}_{ox} + ne^- \leftrightarrow A_{red} \notag\\ B^{n+}_{ox} + ne^- \leftrightarrow B_{red} \end{equation*}\]
These half reactions have reduction potentials \(E_A\) and \(E_B\). Hence, using the Nernst equation and the standard redox potential \(E^\omicron\), we can say that:
\[\begin{equation*} E_A = E^{\omicron}_A - \frac{RT}{nF}\ln\left(\frac{[A_{red}]}{[A^{n+}_{ox}]}\right) \notag\\ E_B = E^{\omicron}_B - \frac{RT}{nF}\ln\left(\frac{[B_{red}]}{[B^{n+}_{ox}]}\right) \end{equation*}\]
Hence, for any two half reactions, we can use equation (5.1):
\[\begin{equation} \Delta E^\omicron = E^\omicron_{\text{electron acceptor}} - E^\omicron_{\text{electron donor}} \tag{5.1} \end{equation}\]
To calculate the free energy for any redox reaction, we can use equation (5.2):
\[\begin{equation} \Delta G^\ = -nF \Delta E^\omicron \tag{5.2} \end{equation}\]
Note that a half equation with a large, positive standard reduction potential has a very high affinity for electrons and is an electron acceptor (i.e., an oxidizing agent). The inverse of the aforementioned statement is also true.
O2 is a strong oxidizing agent, hence its role as the final electron acceptor in the electron transport chain.
5.2.1.1 An example: NADH oxidation
Observe the two half-equations for NADH oxidation:
\[\begin{equation*} NAD^+ + H^+ + 2e^- \leftrightarrow NADH \text{ | } E^{\omicron '} = \text{0.315V} \notag\\ 0.5O_2+ + 2H^+ + 2e^- \leftrightarrow H_2O \text{ | } E^{\omicron '} = \text{0.815V} \end{equation*}\]
Since the O2 / H2O half-equation has the higher \(E^\omicron\), NADH will serve as the electron donor. Hence, the complete equation is:
\[\begin{equation*} 0.5O_2 + NADH + H^+ \leftrightarrow H_2O + NAD^+ \end{equation*}\]
The \(\Delta E^\omicron\) for this reaction is \(0.815 - (-0.315) = 1.130 \text{V}\). Using equation (5.2), the free energy is then:
\[\begin{align} \Delta G^\omicron &= -2(\text{ mol electron / mol reactant}) * 96485 (\text{ C / mol electron}) * 1.13 \text{ J / C} \notag\\ &= -218 \text{ kJ / mol} \end{align}\]
5.2.2 The mitochondrial electron-transport chain
The standard free energy needed to synthesize one mole of ATP from ADP and Pi is 30.5 kJ. Hence, the standard free energy \(\Delta G^\omicron\) is sufficient to drive the formation of several moles of ATP. Like mentioned previously, the electron transport chain allows the standard free energy to be released in smaller, more appropriate quantities - this, when coupled with ATP synthesis, results in a process called oxidative phosphorylation.
The oxidation of one NADH molecule produces 3 molecules of ATP. The thermodynamic efficiency of this reaction is believed to be around 42% under standard conditions, though the thermodynamic efficiency is thought to be around 70% (better than most car engines)!
5.3 Complex I (of the electron transport chain)
This complex harbors one FMN (i.e., flavinmononucleotide) and eight iron-sulfur clusters. Electrons in complex I are transferred from NADH to coenzyme Q via FMN and iron-sulfur clusters in multiple steps.
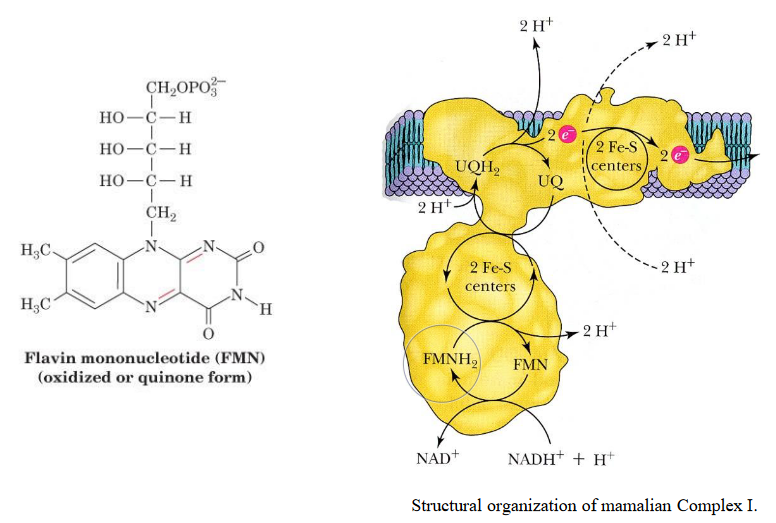
Figure 5.9: Complex I
This complex is about 100 kD (i.e., kiloDaltons) big in eukaryotes and has multiple subunits (as mentioned earlier).
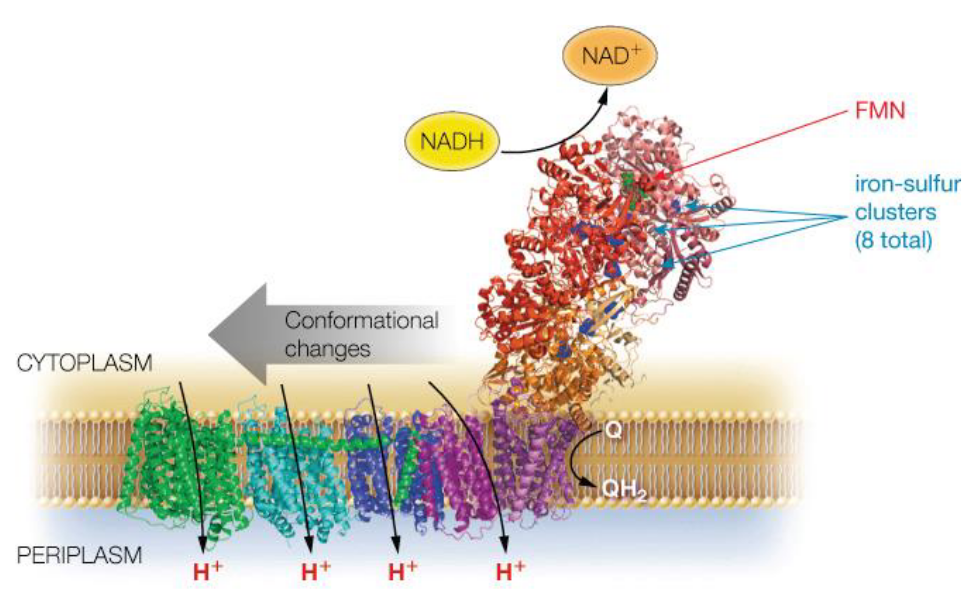
Figure 5.10: Another Visualization of Complex I
It is also important to realize that ubiquinone is a lipophilic electron carrier that is capable of transferring two electrons in a single-electron step.
Ubiquinone has a benzoquinone linked to an isoprene tail:
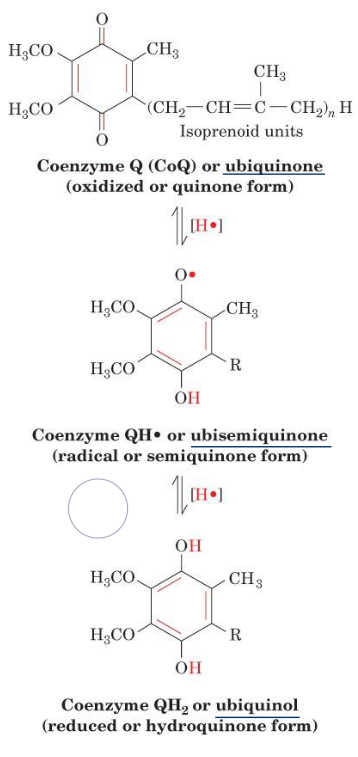
Figure 5.11: Forms of Ubiquinone
Because of this, ubiquinone provides a link between two-electron carriers and single-electron carriers.
5.4 Complex II (again, of the electron transport chain)
Coenzyme Q (see above) can also accept electrons from fatty acid oxidation and from succinate.
Complex II (i.e., succinate dehydrogenase) is located on the inner membranes of mitochondria and is also a part of the Krebs’ cycle. Hence, electrons can be transferred through FAD and a series of iron-sulfur clusters to coenzyme Q:
Figure 5.12: Structure of Complex II
As mentioned previously, complex II does not pump protons into the intermembrane space (remember this)!
5.4.1 Something important about coenzyme Q…
Coenzyme Q collects electrons from flavoproteins (i.e., FMN and FAD), complex I, and complex II and passes them over to complex III.
5.5 Complex III (…of the electron transport chain… again)
This complex catalyzes the transfer of electrons from CoQH2 (i.e., reduced coenzyme Q) to cytochrome C in the intermembrane space. For every electron transferred, two electrons are pumped into the intermembrane space!
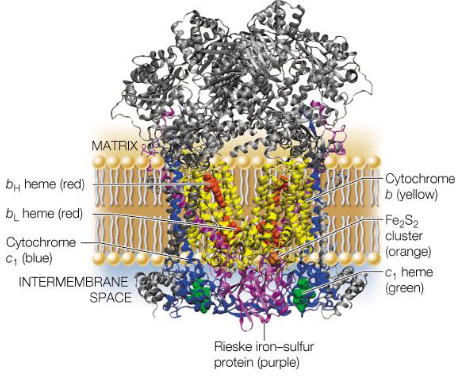
Figure 5.13: Structure of Complex III
Complex III inside mammals is about 250 kD big; each monomer of complex III has about 10 - 11 protein chains.
5.6 The Q Cycle
The Q cycle transfers electrons from two-electron donors over to one-electron donors.
Because each complex III has two Q-binding sites, this requires that electrons from reduced coenzyme Q take two different paths in two stages:
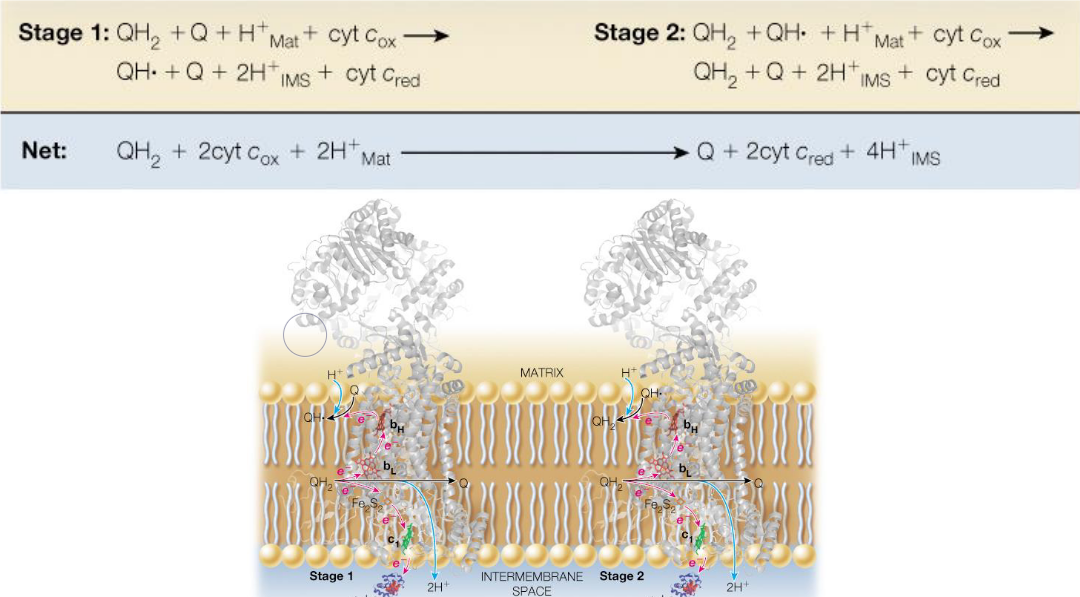
Figure 5.14: Q Cycle Schematic
Four protons are pumped into the intermembrane as a result of the Q cycle.
5.7 Complex IV and Cytochrome C
Mitochondrial cytochrome C has a heme group at the center of its structure:
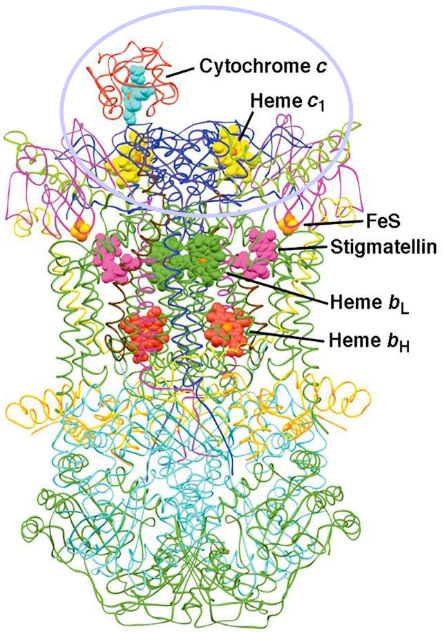
Figure 5.15: Structure of Cytochrome C
Furthermore, cytochrome C is linked to two sulfur atoms. A third sulfur atom from the amino acid methionine engages the iron in cytochrome C.
Cytochrome C - like its coenzyme Q counterpart - is also an electron carrier; it carries electrons from the iron-sulfur clusters of complex III, moves along the mitochondrial membrane surface, and carries electrons to cytochrome C oxidase (i.e. complex IV).
5.7.1 Complex IV
An electron from cytochrome C is first acquired by CuA of the cytochrome C oxidase before it is passed onto Heme a:
Figure 5.16: Redox Sites of Cytochrome C Oxidase
While it is not really understood why the mechanism proceeds as such, it is believed that this is because the heme A-CuA distance is rather (small 11.7 angstroms).
Nevertheless, the electrons are then transferred over to the a3-CuB binuclear center where they are then combined with O2 to form water.
5.8 Outcomes of Electron Transfer
Free energy stored in the proton gradient is called the proton motive force (i.e., the PMF). The PMF has two components:
- The difference in chemical potential energy is denoted by \(\Delta\text{pH}\); this is due to the difference in proton concentrations.
- The electrical potential energy difference is given by \(\Delta\psi\) - this is due to the separation of charge across the membrane.
Hence, the proton motive force is also an electrochemical gradient!
5.8.1 The chemiosmotic theory
In 1961, Scientist Peter Mitchell proposed that the free energy of electron transport is conserved by pumping H+ ions into the mitochondria’s intermembrane space to create a proton motive force (as described above) to generate ATP via F-type ATP synthase.
Figure 5.17: F-Type ATP Synthase
5.8.2 The Boyer mechanism
This mechanism describes the changes in binding in the F1FO subunits of F-type ATP synthase:
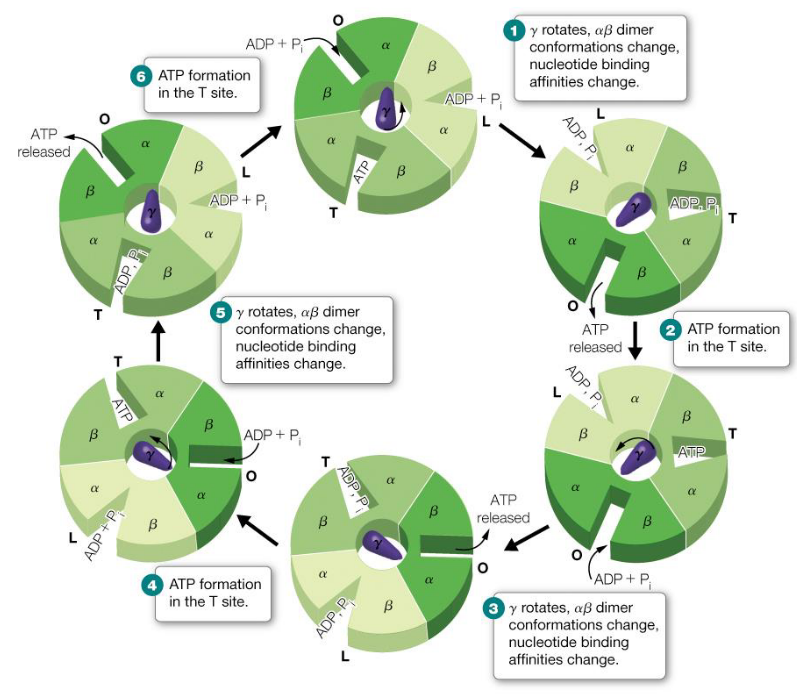
Figure 5.18: Boyer Mechanism
5.8.3 Regulation of ATP synthase through coupling and uncoupling
5.8.3.1 Coupling
As ADP levels in the cell rise, there is an influx of protons and the electrochemical gradient dissapates.
Hence, this stimulates proton pumps in the electron transport chain to pump more protons into the intermembrane space to maintain the proton motive force. More oxygen gas is also consumed as a result.
Furthermore, fuel oxidation pathways (e.g., the TCA cycle) are also stimulated to supply more NADH and FADH2 to the electron transport chain.
5.8.3.2 Uncoupling
Uncouplers dissipate the proton gradient across the inner mitochondrial membrane. Hence, electron transport continues and the latter happens.
However, protons leak back into the mitochondria’s intermembrane space so rapidly that ATP does not occur; this causes the energy released from electron transport to be released as heat instead.
5.9 Brown Fat
Most newborn mammals (us included) are born with brown fat - these adipocyte cells (brown fat are a kind of adipose tissue) have a large amount of mitochondria in them; the mitochondria’s membranes contain a protein called thermogenin. In organisms that live in cold environments, thermogenin makes up 15% of the mitochondria’s inner membrane proteins.
Nevertheless, thermogenin dissipates the proton motive force by allowing protons to return to the mitochondrial matrix, hence allowing newborn animals to maintain their body temperature.
Skunk cabbage and other related species contain floral spikes that also use a similar mechanism to evaporate odiferous molecules to attract insects to fertilize them.
In brown fat, norepinephrine can be used to uncouple oxidative phosphorylation:
Figure 5.19: Uncoupling of Oxidative Phosphorylation in Brown Fat Mitochondria