2 Data Generation
2.2 Pileup
SCISSOR package1 was applied to generate pileup from BAM file, region info, and coverage plots based on its tutorial.
A comprehensive gene annotation file (GTF) can be downloaded at GENCODE.
From the GTF file, build_gaf function creates a region info file named SCISSOR_gaf.txt and shows the full path of the file. The file contains regions by gene_name and gene_id.
For instance, we can make pileup and compare coverage plots of part_intron and only_exon using gene KEAP1 and 5 samples.
If 5 bam files are saved in the data/bamfiles folder after Samtools, the full file path of them can be set as BAMfiles.
gen_pileup function saves pileup, regions, and Ranges for Gene, which become inputs to plot pileup figures.
BAMfiles <- list.files(path=paste0("./data/bamfiles"), pattern="\\.sort.bam$", full.names=TRUE)
caseIDs <- substr(basename(BAMfiles), start=6, stop=22)
par(mfrow=c(1,2))
# part_intron
gen_pileup(Gene="KEAP1",
regionsFile=data.table::fread(file="./data/SCISSOR_gaf.txt"),
BAMfiles=BAMfiles,
caseIDs=caseIDs,
outputdir=paste0("./output/readBAM/"))
load(paste0("./output/readBAM/KEAP1_pileup_part_intron.RData"))
SCISSOR::plot_pileup(Pileup=log10(pileup+1), case=caseIDs, Ranges=Ranges, logcount=1, main="part_intron")
# only_exon
pileupData = SCISSOR::build_pileup(Pileup=pileup, case=caseIDs, regions=regions, inputType="part_intron", outputType="only_exon")
geneRanges = SCISSOR::get_Ranges(Gene=Ranges$Gene, regions=regions, outputType="only_exon")
SCISSOR::plot_pileup(Pileup=log10(pileupData+1), case=caseIDs, Ranges=geneRanges, logcount=1, main="only_exon")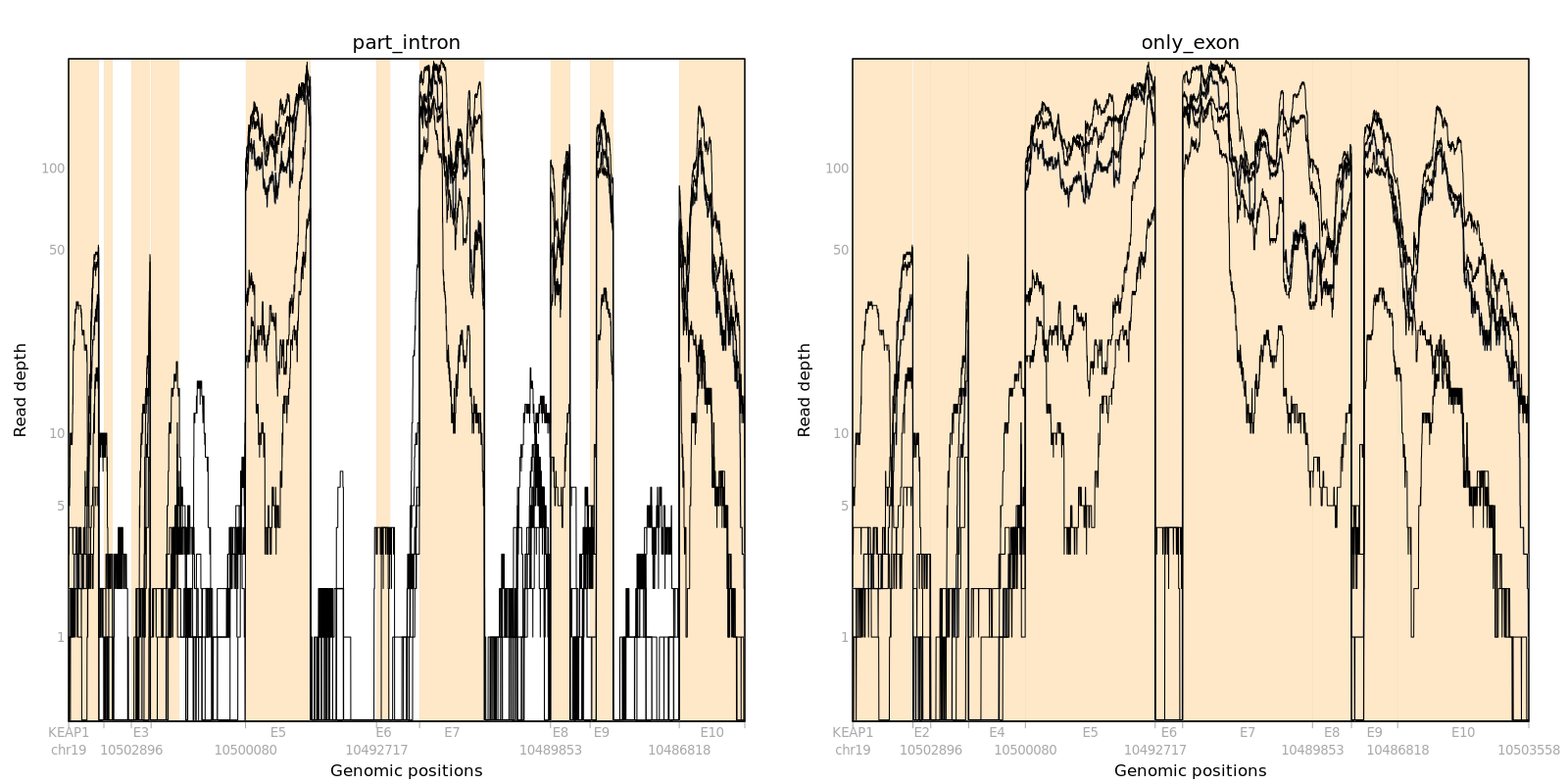
## ip.start e.start e.end ip.end ip.start e.start e.end ip.end
## exon1 10503558 10503558 10503241 10503187 10503558 10503558 10503241 10503241
## exon2 10503186 10503186 10503093 10502897 10503186 10503186 10503093 10503093
## exon3 10502896 10502896 10502697 10502688 10502896 10502896 10502697 10502697
## exon4 10502687 10502687 10502388 10502039 10502687 10502687 10502388 10502388
## exon5 10500429 10500080 10499395 10499046 10500080 10500080 10499395 10499395
## exon6 10493066 10492717 10492573 10492263 10492717 10492717 10492573 10492573
## exon7 10492262 10492262 10491577 10491228 10492262 10492262 10491577 10491577
## exon8 10490202 10489853 10489648 10489436 10489853 10489853 10489648 10489648
## exon9 10489435 10489435 10489192 10488843 10489435 10489435 10489192 10489192
## exon10 10487167 10486818 10486125 10486125 10486818 10486818 10486125 10486125Choi, H.Y., Jo, H., Zhao, X. et al. SCISSOR: a framework for identifying structural changes in RNA transcripts. Nat Commun 12, 286 (2021). https://doi.org/10.1038/s41467-020-20593-3↩︎